From: Chapter 16, The BLAST Sequence Analysis Tool

NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
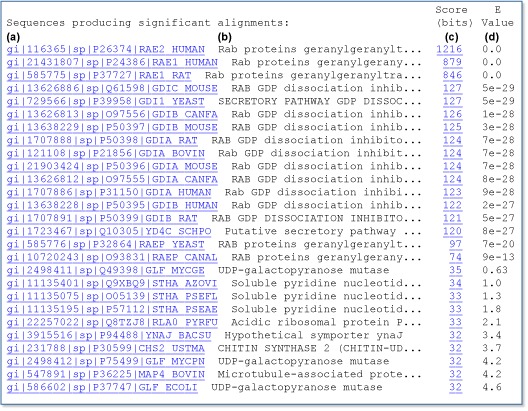
Each line is composed of four fields: (a) the gi number, database designation, Accession number, and locus name for the matched sequence, separated by vertical bars (Appendix 1); (b) a brief textual description of the sequence, the definition. This usually includes information on the organism from which the sequence was derived, the type of sequence (e.g., mRNA or DNA), and some information about function or phenotype. The definition line is often truncated in the one-line descriptions to keep the display compact; (c) the alignment score in bits. Higher scoring hits are found at the top of the list; and (d) the E-value, which provides an estimate of statistical significance. For the first hit in the list, the gi number is 116365, the database designation is sp (for SWISS-PROT), the Accession number is P26374, the locus name is RAE2_HUMAN, the definition line is Rab proteins, the score is 1216, and the E-value is 0.0. Note that the first 17 hits have very low E-values (much less than 1) and are either RAB proteins or GDP dissociation inhibitors. The other database matches have much higher E-values, 0.5 and above, which means that these sequences may have been matched by chance alone.
From: Chapter 16, The BLAST Sequence Analysis Tool

NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.