Clinical Description
Kagami-Ogata syndrome is characterized by developmental delay, intellectual disability, feeding difficulty, full cheeks and prominent and deep philtrum, small bell-shaped thorax with coat-hanger appearance of the ribs, and abdominal wall defects (omphalocele and diastasis recti). Additional common features include joint contractures, kyphoscoliosis, coxa valga, and laryngomalacia. Cardiac disease and hepatoblastoma have also been reported [Kagami et al 2015, Ogata & Kagami 2016]. To date, approximately 100 individuals have been diagnosed with Kagami-Ogata syndrome [Sakaria et al 2021; Mackay et al 2022; Smith et al 2024; T Ogata & M Kagami, unpublished observations].
Table 2.
Kagami-Ogata Syndrome: Frequency of Select Features
View in own window
| Feature | % of Persons w/Feature 1 | Comment |
|---|
|
Pregnancy & delivery
| Polyhydramnios | >95% | |
| Placentomegaly | ~85% | Placenta size >120% of normal |
|
Development & cognition
| Developmental delay | >95% | Moderate to severe |
| Intellectual disability | 100% | |
|
Nutrition & growth
| Feeding difficulty | >95% | |
| Prenatal overgrowth | >50% | Birth length &/or weight >2 SD above mean |
| Postnatal growth deficiency | ~35% | Height &/or weight >2 SD below mean |
|
Craniofacial features
| Full cheeks & prominent & deep philtrum (most common & specific features) | >90%-95% | Additional features incl frontal bossing, hirsute forehead, blepharophimosis, depressed nasal bridge, anteverted nares, puckered (somewhat narrow & tented) lips, micrognathia, & short, webbed neck |
|
Skeletal abnormalities
| Small bell-shaped thorax | 100% | |
| Coat-hanger appearance of ribs | 100% | |
| Joint contractures | 60%-65% | |
| Kyphoscoliosis | ~40% | |
| Coxa valga | ~33% | |
|
Respiratory
| Laryngomalacia | ~40% | |
|
Abdominal wall defects
| Diastasis recti | 65%-70% | |
| Omphalocele | ~30% | |
|
Other features
| Cardiac disease | 25% | |
| Hepatoblastoma | 5%-10% | |
Pregnancy and delivery. Polyhydramnios is typically identified in the second trimester at a median gestational age of 25.5 weeks (range: 14-30 weeks' gestation); amnioreduction is required in most pregnancies after 25 weeks' gestation (~80%) and in almost all pregnancies after 30 weeks' gestation. Polyhydramnios is due to placentomegaly and impaired swallowing. Thoracic and abdominal abnormalities are found on prenatal ultrasound in ~40% of fetuses from approximately 25 weeks' gestation. Premature delivery is frequent (~80%), with a median gestational age at delivery of 32.5 weeks (range: 30-35 weeks' gestation).
Developmental delay. Head control occurs at a median age of seven months (range: 3-36 months). Sitting without support occurs at a median age of 12 months (range: 8-25 months). Walking without support occurs at a median age of 25.5 months (range: 20-49 months). Only mild delay has been reported in two individuals with Kagami-Ogata syndrome: one individual with an epimutation [Higashiyama et al 2022] and one with mosaic paternal uniparental disomy of chromosome 14 (upd(14)pat) [Haug et al 2017].
Intellectual disability. Intellectual disability is reported in all individuals, with a median IQ of 55 (range: 29-70).
No abnormal brain MRI findings were delineated in five individuals examined.
Growth. Prenatal growth is well preserved and birth weight and length are usually above the mean. Range in birth weight is 0.1 standard deviations (SD) below the mean to 8.8 SD above the mean, and range in birth length is 1.7 SD below the mean to 3.0 SD above the mean. However, postnatal growth is frequently compromised primarily due to poor nutrition related to respiratory failure and feeding difficulty. In individuals age one to 15 years, range in weight is 6.0 SD below the mean to 4.0 SD above the mean, and range in length/height is 8.7 SD below the mean to 1.1 SD above the mean.
Craniofacial features (see ) in individuals with Kagami-Ogata syndrome include frontal bossing (~75%), hirsute forehead (~70%), blepharophimosis (70%-75%), full cheeks (>90%), depressed nasal bridge (>90%), anteverted nares (80%), prominent and deep philtrum (>95%), puckered (somewhat narrow and tented) lips (>50%), micrognathia (~100%), and short, webbed neck (>90%). Craniofacial features are typically present in infancy and consistently recognizable during childhood. While many of the craniofacial features are nonspecific, full cheeks and prominent and deep philtrum are very common and appear specific to Kagami-Ogata syndrome.
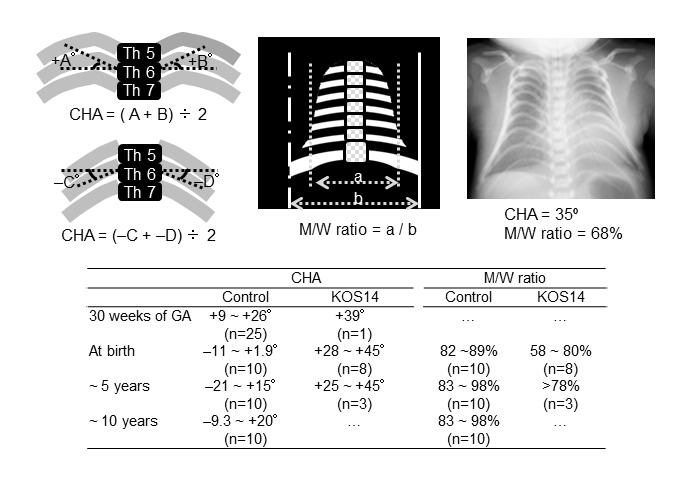
Thoracic abnormalities with respiratory failure. A small bell-shaped thorax is evident in infancy. Objective assessment of increased coat-hanger angle and decreased mid-to-widest thorax ratio is possible from fetal life through childhood (see ). The shape of the thorax appears normal after infancy, whereas coat-hanger appearance of the ribs remains discernible throughout childhood. Because of the thoracic anomalies, mechanical ventilation with oxygen has been required in more than 90% of individuals, with a median duration of one month (range: 0.1-17 months). Tracheostomy is sometimes necessary due to severe laryngomalacia.
Other skeletal abnormalities. Contractures at various joints, kyphoscoliosis, and coxa valga are also reported. The severity of these findings is variable among affected individuals.
Feeding/gastrointestinal manifestations. Tube feeding was required in all individuals who survived more than one week, with a median duration of 7.5 months (range: 0.1-89 months). Diastasis recti is common, and omphalocele can also occur. Abdominal wall weakness may cause constipation.
Cardiac disease. Cardiovascular malformations are recognized in ~25% of affected individuals. Of eight individuals who have been confirmed to have cardiac malformations, two have atrial septal defect, one has ventricular septal defect, four have patent ductus arteriosus, and one has pulmonary stenosis. These cardiac lesions are mild and do not require intensive medication or surgery. No individuals have been reported to have cardiomyopathy or conduction defects.
Hepatoblastoma has been identified in three individuals during infancy out of the 100 reported individuals to date.
Other. Simple seizure was reported in one individual [Kagami et al 2015].
Prognosis. Mortality is relatively high before age five years (20%-25% of individuals), but increased mortality is not observed in those with Kagami-Ogata syndrome older than age five years. The cause of death is variable; respiratory failure constitutes the primary cause of death. To date, nine adults age 18 years and older with Kagami-Ogata syndrome have been identified, including four unpublished individuals; the oldest affected individual is age 35 years [Smith et al 2024; T Ogata & M Kagami, personal observations].