NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Polyomaviruses of the BK- and JC-strains often remain latent within the transitional cell layer of the bladder, ureters and the renal pelvis as well as in tubular epithelial cells of the kidney. Slight changes in the immune status and/or an immunocompromised condition can lead to the (re)activation of latent polyomaviruses, especially along the transitional cell layer, resulting in the shedding of viral particles and infected cells into the urine. A morphologic sign of the (re)activation of polyomaviruses is the detection of typical intranuclear viral inclusion bearing epithelial cells, so-called “decoy cells”, in the urine. Decoy cells often contain polyoma-BK-viruses. The inclusion bearing cells are easily identified and quantifiable in routine Papanicolaou stained urine cytology specimens. With some experience, decoy cells can also be detected in the unstained urinary sediment by phase contrast microscopy. Different morphologic variants of decoy cells (types 1 through 4) are described and ancillary techniques (immunohistochemistry, electron microscopy (EM), and fluorescence-in-situ-hybridization (FISH)) for proper identification and characterization are discussed. Special emphasis is placed on the clinical significance of the detection of decoy cells as a parameter to assess the risk for disease, i.e., polyoma-BK-virus nephropathy (BKN) in kidney transplant recipients. The sensitivity and specificity of decoy cells for diagnosing BKN is 99% and 95%, respectively, the positive predictive value varies between 27% and more than 90%, and the negative predictive value is 99%. The detection of decoy cells is compared to other techniques applicable to assess the activation of polyomaviruses in the urine (polymerase chain reaction (PCR) and EM).
Introduction
General aspects of polyomaviruses are discussed in detail elsewhere in this handbook. Here, it is important to emphasize that polyomaviruses are often not cleared from the body after the primary infection. Rather, it is assumed that primary viral entry into the host, often via an upper respiratory infection, results in transient viremia and viral spread to permissive tissues, in particular, to transitional cells and renal tubular epithelial cells. Polyomaviruses can establish life-long latency under normal cellular and humoral immuno-surveillance.1-3 Latent polyomavirus infections cannot be identified histologically or immunohistochemically but rather require the use of molecular techniques for detection (Southern blot or PCR analyses).1,2 Disease caused by (latent) polyomaviruses is typically not seen in the immunocompetent host. However, even slight changes in the immune surveillance can result in transient, asymptomatic and self-limiting activation of latent polyomaviruses in healthy individuals. Since the urothelium is a common site of viral latency, reactivation of polyomaviruses often occurs in the transitional cell layer. Such viral (re)activation is characterized by the shedding of viral particles and viral inclusion bearing epithelial cells (so-called “decoy cells”) into the urine. Indeed, the first strain of human polyomaviruses was isolated from the urine in 1971 and named “BK-polyomavirus” strain after the initials of the patient.4 Based on the detection of decoy cells in the urine, transient and asymptomatic reactivation of polyomaviruses can be seen in 0.5-0.6% of all urine cytology specimens.5,6 A high prevalence of decoy cell shedding is found in pregnant women (3%), patients suffering from cancer (13%), and diabetes mellitus (3%), as well as in healthy renal allograft (23%) and pancreas transplant (11%) recipients.6-13 Decoy cell shedding has also been reported after heart transplantation.14 Polyomavirus (re)activation and the shedding of decoy cells are generally not associated with kidney dysfunction, i.e., a rise in serum creatinine levels, or other renal abnormalities.8,13
In contrast, in severely immunocompromised patients, polyomaviruses can cause manifest disease. With the advent of new, highly potent immunosuppressive drug regimens introduced into the management of renal transplant recipients, the activation and replication of polyomaviruses of the BK-strain in renal tubules of the allograft, i.e., polyoma-BK-virus allograft nephropathy (BKN), has gained great clinical significance. BKN is characteristically associated with signs of viral activation, i.e., the shedding of decoy cells. Decoy cells contain mostly BK-virus antigens. Thus, in renal allograft recipients, the examination of urine cytology specimens and the search for polyomavirus inclusion bearing cells can be used as a clinical tool to assess the (re)activation of latent polyomaviruses and the risk for BKN (see below). In bone marrow transplant recipients, massive replication of BK-virus in the bladder mucosa and the shedding of decoy cells are associated with a hemorrhagic cystitis several weeks post grafting. However, BKN is not seen after bone marrow transplantation. A productive infection with the polyoma-JC-virus strain in the brain (i.e., in oligodendrocytes) of Acquired Immune Deficiency syndrome (AIDS) patients can cause “progressive multifocal leukoencephalopathy” which is generally not associated with renal or urinary abnormalities.
In the following paragraphs, we will characterize polyomavirus inclusion bearing “decoy-cells”. We will emphasize the clinical significance of decoy cells for assessing the risk of BKN in kidney transplant recipients. The morphological detection of decoy cells will be compared with other ancillary techniques, such as PCR analyses, electron microscopy, and FISH analyses.
Polyomavirus Inclusion Bearing “Decoy” Cells
Beginning in 1945, George Papanicolaou stressed the usefulness of urine cytology examination and the “Papanicolaou stain” for the diagnostic evaluation of cellular elements in voided urine specimens. This technique rapidly gained worldwide acceptance since it provided an easy, reliable, and inexpensive clinical tool. Approximately forty years ago, Koss and colleagues described polyomavirus inclusion bearing cells for the first time in urine cytology specimens.15 They coined the term “decoy cells” to alert pathologists not to misdiagnose viral inclusion bearing cells as malignant cancer cells.
Decoy-Cells, Morphology and Characterization
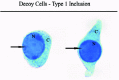
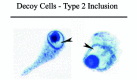
The name “decoy cell” is a descriptive term for epithelial cells with intranuclear viral inclusion bodies that can have different phenotypes (types 1-4) depending upon the state of viral replication and maturation as well as the state of cellular preservation. The order in which the various phenotypes may occur during intranuclear viral assembly is unclear. Hybrid forms representing transitions between the different phenotypes are frequently found in the same specimen. Most common are classic decoy cells characterized by large, homogenous, amorphous ground-glass like intranuclear inclusion bodies and a condensed rim of chromatin (type 1) (fig. 1). Sometimes, decoy cells reveal granular intranuclear inclusions surrounded by a clear halo, i.e., cytomegalovirus (CMV)-like (type 2) (fig. 2). Occasionally, multinucleated decoy cells with granular chromatin are detected (type 3) (fig. 3). Type 4 decoy cells show vesicular nuclei, often with clumped chromatin and nucleoli (fig. 4). Koss called these latter inclusions the “empty post-inclusion stage”.15 Types 3 and 4 decoy cells are especially prone to misinterpretation as “cancer cells”. Although the nuclear features are most characteristic, many decoy cells additionally show a typical eccentric cytoplasm resembling the tail of a comet (termed “comet cells” by some, fig. 1 and fig.2).16
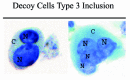
Figure 3
Type 3 decoy cells showing a granular chromatin pattern and multinucleation (N), cytoplasm (C). Papanicolaou stained preparations, 400 x original magnification.

Figure 4
Type 4 decoy cells with vesicular nuclei and a distinct network of coarsely granular and clumped chromatin. Papanicolaou stained preparations, 400 x original magnification.
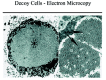
Decoy cells mostly contain polyomaviruses of the BK strain or less commonly of the JC strain. Rarely, also adenoviruses may be found (Table 1). 5,17,18 Immunohistochemical and electron microscopical analyses can easily be used to verify the presence of viruses and to identify the virus families. In general, most types 1 through 4 intranuclear inclusion bodies in decoy cells give a positive staining reaction with a commercially available antibody detecting the simian virus “SV-40 T antigen” which is common to all known polyomavirus strains pathogenic in humans (i.e., BK-, JC-, SV-40 strains (fig. 5); see appendix for staining protocols).7,19 Of note: since the large T antigen is only expressed in abundancy during the early phases of viral replication, decoy cells with late stages of polyomavirus assembly may be “T antigen” negative. Using BK-virus specific antibodies or PCR techniques, most decoy cells contain polyoma-BK-virus particles. 5 Immunohistochemistry can also help to identify adenovirus containing decoy cells. Electron microscopy is well suited to detect polyomaviruses and adenoviruses based on their characteristic size of 40—50 nm and 80 nm, respectively (fig. 6). Ultrastructural analysis, however, is not suited to distinguish between different polyoma- or adenovirus strains. Although productive infections with cytomegalovirus, herpes simplex virus or human papillomavirus can show nuclear abnormalities including viral inclusion bodies, typical “decoy cells” as described above are generally not found in the urine in these infections (Table 1).
Table 1
Cytological changes induced by the most common viral infections observed in urine cytology specimens.
Decoy-Cells, Origin
The origin of decoy cells cannot be easily discerned based on morphologic grounds. It seems likely to us that they would commonly originate from the urothelium, in particular, in healthy and asymptomatic patients (fig. 7a).5,7 This assumption is based on the observation that the urothelium often harbors latent BK-virus infections (approximately 50% of individuals, personal observation). The replication of polyomaviruses is most pronounced in the superficial transitional cell layer, i.e., in umbrella cells, which can easily be shed. 9 As mentioned above, decoy cell shedding is often asymptomatic and renal function remains unaltered. Polyomavirus inclusion bearing cells are never seen in native kidneys of immune competent patients further arguing for an extra (renal) parenchymal origin of decoy cells found in the urine of healthy individuals.
In contrast, in immunocompromised patients BKN is characterized by intra-renal replication of BK viruses and kidney dysfunction. The morphological signs of viral replication in renal tubular epithelial cells in cases of BKN are very similar to those seen in transitional cells and decoy cells (see Chapter 12: “Latent and Productive Polyomavirus Infections of Renal Allografts: Morphological, Clinical and Pathophysiological Aspects”). Thus, in cases of BKN, decoy cells likely also originate from the renal parenchyma.7,9,19 It is tempting to speculate that BKN may be caused by an ascending route of infection with spreading of polyomavirus replication from transitional cells to collecting ducts and proximal tubular epithelial cells in some patients in whom risk factors provide the right window of opportunity (fig. 7b and fig. 7c).7 However, this hypopthesis has not yet been proven.
Decoy-Cells Versus Malignant Tumor Cells
One of the most important challenges, already stressed by Koss and his colleagues, is to properly identify decoy cells and to avoid their misinterpretation as “malignant tumor cells”. 15Sound knowledge of the various phenotypes of viral inclusion bodies and the utilization of immunohistochemical and electron microscopic analyses should generally lead to their proper identification. In cases of polyomavirus activation and replication, the evaluation of “atypical cells” with proliferation markers or by DNA image cytometry can be misleading. Polyomaviruses require the “machinery” of the host cells for viral amplification. Thus, immunohistochemical stains to detect “proliferation associated antigens”, such as antibodies directed against proliferating cell nuclear antigen (PCNA), KI-67 or MIB-1, give strong signals in decoy cells and inclusion bearing transitional cells. Such staining profiles should not be misinterpreted as a sign of marked “cell ” proliferation, but rather indicate the replication of viral DNA. Accordingly, DNA cytometry/histograms of decoy cells invariably show aneuploidy due to the viral DNA content (fig. 8). In contrast, FISH with chromosome enumeration probes and single locus specific identifiers (9p21) (UroVision,TM Vysis Inc., Downers Grove IL), can reliably demonstrate normal chromosome and gene copy numbers (fig.8). 20 The FISH profile clearly identifies decoy cells as “benign” and distinguishes them from cancer cells.21
Decoy-Cells, Detection
Decoy cells can best be identified and quantified in standard alcohol fixed and Papanicolaou stained urine cytology specimens from either smeared or cytocentrifuged (i.e., cytospin) urine samples. In addition, the recently introduced monolayer technique is also feasible since it provides excellent preservation of nuclear details (Cytyc Inc., Boxborough, MA, and Tripath Imaging Inc., Burlington, NC). The second morning midstream voided urine specimen is best suited as the high level of cellular degeneration severely limits the first morning specimen. A few important considerations in the handling of urine specimens are the following: (1) Fresh urine specimens should be promptly transported to the cytology laboratory for immediate processing. (2) An alternative and more frequently used method is the fixation of the urine with an equal volume of 50-70% ethyl alcohol, preferably with added 2% Carbowax (polyethylene glycol). This procedure can be performed beside or in the laboratory when delayed specimen handling is anticipated. (3) Conventional cytospin or smear preparations should be obtained immediately after the specimen is received in the cytology laboratory. (4) The use of coated glass slides is recommended for adequate adherence of the cellular and noncellular elements to the slide surface. 22 It should be noted that specific guidelines for adequacy of voided urine specimens at the time of cytologic interpretation have not been firmly established and much is still dependent on the experience level of the pathologist. 22 Decoy cells can also be detected in the unstained urine sediment using phase contrast microscopy.8,23 This technique, however, requires great experience and the quantification of decoy cells is tricky. We, therefore, recommend the analysis of standard Papanicolaou stained cytology specimens as described above.
Urine Analysis for Risk Assessment and Management of BK-Virus Nephropathy (BKN)
BKN is the most important infectious complication affecting renal allografts with a reported prevalence of 1% to 10%. It is typically caused by the replication of BK-virus in tubular epithelial cells, hence the name. The polyoma-JC and SV40 virus strains are only rarely (co)activated. BKN often results in chronic allograft dysfunction or even loss. The definitive diagnosis of BKN can only be made histologically in a renal allograft biopsy specimen showing characteristic tubular changes (see Chapter 12 “Latent and Productive Polyomavirus Infections of Renal Allografts: Morphological, Clinical and Pathophysiological Aspects”).7,9-11,19,24,25 Depending on the histologic stage in which BKN is first diagnosed, the outcome may vary. Best clinical results are seen if BKN is detected early (histological stage/pattern A), at a time when graft function is largely unaltered and irreversible graft fibrosis and tubular atrophy are absent.7,10,19,24,26-28 Such an early diagnosis requires: (a) proper risk assessment of renal allograft recipients, and (b) optimal timing of a renal allograft biopsy. The search for decoy cells in the urine can assist in achieving these goals.7,9--11,19,24,30 Clinical risk assessment strategies, including the search for decoy cells, were extensively discussed at the first “Polyomavirus Allograft Nephropathy Consensus Conference” held in Basel, Switzerland in October 2003 (manuscript in preparation).
Urine Cytology
As outlined above, the shedding of decoy cells generally indicates the (re)activation of (BK) polyomaviruses in the urothelium. Such (re)activation is a prerequisite for the potential development of BKN if the right “window of opportunity” for unrestricted viral replication in tubular epithelial cells is provided.7,10,19 We retrospectively analyzed urine samples from more than 300 renal allograft recipients and found decoy cell shedding in 23% of patients; in 7% in high numbers, i.e., more than 5 decoy cells per 10 high power microscopic fields in cytology smears or alternatively more than 10 decoy cells per cytospin preparation.7,11 BKN was diagnosed in 2% of patients, all of whom demonstrated abundant decoy cell shedding which often preceded the histological diagnosis of BKN by weeks to months.7,9,10,19 These observations were confirmed in a prospective analysis.31 In our hands, the detection of high numbers of decoy cells had a positive predictive value to indicate BKN of 27% and a negative predictive value of 99%, i.e., “no decoy cells, no BKN”.7,10 The positive predictive value can be further increased to over 90% by taking additional parameters into consideration: (a) a “dirty” cytological background, (b) decoy cell shedding in the setting of allograft dysfunction, (c) extended and persistent decoy cell shedding over more than 6 weeks, and (d) the detection of decoy cell casts.11,19,24 The latter finding is considered to be pathognomonic for BKN since “cast material” always originates from the kidney parenchyma, i.e., the renal tubular compartment. In histologically confirmed cases of BKN, the number of decoy cells correlates with the number of inclusion bearing renal tubular epithelial cells. 24 Thus, the detection of decoy cells can also be used during therapeutic attempts to monitor for decreased viral loads and ultimately for viral “clearance” from the transplanted kidney.7,10,19,24,31 Clinically, the search for decoy cells in the urine is frequently supplemented by (quantitative) PCR studies of BK virus DNA loads in plasma samples which can provide very valuable additional clinical information (fig. 7c).10,29,31-33
Ancillary Techniques
Besides the search for decoy cells, PCR and EM analyses of urine samples have also been used to evaluate the activation of polyomaviruses and to assist with patient management.32,34-36 All tests can provide important information, however, they vary greatly in sensitivity, specificity, feasibility, time requirement, and cost. So far, exhaustive comparative analyses have not been performed. A test should be carefully chosen in order to address specific questions.
Electron microscopy (EM) of negatively stained urine samples can be easily used to rapidly identify polyomaviruses in large numbers.34,36 Howell and colleagues studied six patients with BKN, all of whom showed icosahedral, nonenveloped polyomavirus particles in the urine. 34 In three patients in whom BK virus replication in the kidney ceased during follow-up, i.e., BKN “cleared from the graft”, viral particles also disappeared from the urine. The investigators did not find polyomaviruses in control patients (potentially due to low copy numbers below the level of detection, which varies between 10 3 and 10 9 viral particles per ml urine). 36 Thus, EM of negatively stained urine specimens provides an additional rapid, noninvasive, and relatively inexpensive diagnostic tool for the detection of large numbers of polyomaviruses. Such “crude” analyses appear to be suitable for patient management risk assessment.
In contrast, highly sensitive PCR studies of urine samples do not seem to be of great clinical benefit in the setting of kidney or bone marrow transplantation since PCR tests often detect clinically irrelevant (low) levels of BK virus activation, i.e., low positive predictive value to indicate disease. In addition, urine PCR analyses can be technically challenging, due to nonspecific endogenous inhibition of the PCR reactions or cross-contamination problems. Whether quantitative PCR tests measuring BK virus DNA or RNA loads in urine samples may be better suited for patient management has to be determined in future multicenter studies.32,35 Not surprisingly, one report suggests that viral load levels in the urine have to be very high and exceed 107 BK virus DNA copies per ml in order to be predictive of BKN.32
From a clinical point of view, we propose a step-wise approach to assess the risk of renal allograft recipients for BKN. Initially (step one),patients should be screened for the activation of BK-virus. As outlined above, this goal can most easily be achieved by searching in the urine for decoy cells or alternatively for viral particles by EM. Positive test results should further be amended by quantitative PCR analyses measuring BK virus DNA loads in serum samples (step two). SERUM BK-virus load levels exceeding 10,000 copies/ml indicate a very high risk for BKN.31This algorithmic approach will help to properly identify kidney transplant recipients in whom a diagnostic graft biopsy should be performed7,10,19,29,31,33 This concept has largely been adopted by the “Polyomavirus Allograft Nephropathy Consensus Conference” (Basel, Switzerland, October 2003, manuscript in preparation).
Appendix Immunohistochemical Staining Protocols to Detect Polyomavirus Antigens in Urine Cytology Specimens (Decoy Cells)
In order to detect polyomavirus antigens in decoy cells, we generally use a monoclonal antibody directed against the large T antigen of the SV-40 polyomavirus strain (Oncogene Research Products, San Diego, CA, USA, Cat #DP02, DP02A, clone PAB 416). This antibody typically detects the T antigen of the BK-, JC-, and SV-40 strains (i.e., “pan” anti-polyomavirus antibody). Thus, the immunohistochemical detection of the SV40-T antigen can only prove the presence of polyomavirus antigens; different polyomavirus strains cannot be differentiated. The antibody typically gives a crisp nuclear staining reaction in some, but not all decoy cells (likely due to different stages of viral assembly and maturation since the large T antigen is expressed during early viral replication).
BK-viruses can be detected with a monoclonal antibody specifically directed against the T region of the polyoma-BK-virus strain (Chemicon, Mab8505, clone BK-T1). This antibody does not cross-react with JC-viruses. The antibody often shows increased background staining.
For all cytology specimens, we use antigen retrieval by microwaving for 5 minutes at 80 degrees Celsius followed by overnight incubation with the primary antibody at a dilution of 1:20.000 at 4 degrees Celsius. Subsequent to the incubation with a secondary antibody AEC is used as a chromogen. Histological sections of known cases of BKN can serve as positive staining controls.
References
- 1.
- Nickeleit V, Singh HK, Gilliland MGF. et al. Latent polyomavirus type BK loads in native kidneys analyzed by Taqman PCR: What can be learned to better understand BK virus nephropathy? J Am Soc Nephrol. 2003;14:42–4A.
- 2.
- Chesters PM, Heritage J, McCance DJ. Persistence of DNA sequences of BK virus and JC virus in normal human tissues and in diseased tissues. J Infect Dis. 1983;147(4):676–684. [PubMed: 6302172]
- 3.
- Heritage J, Chesters PM, McCance DJ. The persistence papovavirus BK DNA sequences in normal human renal tissue. J Med Virol. 1981;8:143–150. [PubMed: 6271922]
- 4.
- Gardner SD, Field AM, Coleman DV. et al. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1:1253–1257. [PubMed: 4104714]
- 5.
- Itoh S, Irie K, Nakamura Y. et al. Cytologic and genetic study of polyomavirus-infected or polyomavirus-activated cells in human urine. Arch Pathol Lab Med. 1998;122(4):333–337. [PubMed: 9648901]
- 6.
- Kahan AV, Coleman DV, Koss LG. Activation of human polyomavirus infection-detection by cytologic technics. Am J Clin Pathol. 1980;74(3):326–332. [PubMed: 6251715]
- 7.
- Nickeleit V, Hirsch HH, Zeiler M. et al. BK-virus nephropathy in renal transplants-tubular necrosis, MHC-class II expression and rejection in a puzzling game. Nephrol Dial Transplant. 2000;15(3):324–332. [PubMed: 10692517]
- 8.
- Binet I, Nickeleit V, Hirsch HH. et al. Polyomavirus disease under new immunosuppressive drugs: A cause of renal graft dysfunction and graft loss. Transplantation. 1999;67(6):918–922. [PubMed: 10199744]
- 9.
- Nickeleit V, Hirsch HH, Binet IF. et al. Polyomavirus infection of renal allograft recipients: From latent infection to manifest disease. J Am Soc Nephrol. 1999;10(5):1080–1089. [PubMed: 10232695]
- 10.
- Nickeleit V, Singh HK, Mihatsch MJ. Polyomavirus nephropathy: Morphology, pathophysiology, and clinical management. Curr Opin Nephrol Hypertens. 2003;12:599–605. [PubMed: 14564196]
- 11.
- Drachenberg CB, Beskow CO, Cangro CB. et al. Human polyoma virus in renal allograft biopsies: Morphological findings and correlation with urine cytology. Hum Pathol. 1999;30(8):970–977. [PubMed: 10452511]
- 12.
- Hogan TF, Padgett BL, Walker DL. et al. Survey of human polyomavirus (JCV, BKV) infections in 139 patients with lung cancer, breast cancer, melanoma, or lymphoma. Prog Clin Biol Res. 1983;105:311–324. [PubMed: 6304768]
- 13.
- Haririan A, Hamze O, Drachenberg CB. et al. Polyomavirus reactivation in native kidneys of pancreas alone allograft recipients. Transplantation. 2003;75(8):1186–1190. [PubMed: 12717201]
- 14.
- Masuda K, Akutagawa K, Yutani C. et al. Persistent infection with human polyomavirus revealed by urinary cytology in a patient with heart transplantation, a case report. Acta Cytol. 1998;42(3):803–806. [PubMed: 9622713]
- 15.
- Koss LG. The urinary tract in the absence of cancerIn: Koss LG, ed.Diagnostic Cytology and Its Histopathologic Basis4th ed. Philadelphia: J.B. Lippincott19922890–933.
- 16.
- Crabbe JG. “Comet” or “decoy” cells found in urinary sediment smears. Acta Cytol. 1971;15(3):303–305. [PubMed: 4104212]
- 17.
- De Las Casas LE, Hoerl HD, Bardales RH. et al. Utility of urinary cytology for diagnosing human polyoma virus infection in transplant recipients: A study of 37 cases with electron microscopic analysis. Diagn Cytopathol. 2001;25(6):376–381. [PubMed: 11747234]
- 18.
- Asim M, Chong-Lopez A, Nickeleit V. Adenovirus infection of a renal allograft. Am J Kidney Dis. 2003;41(3):696–701. [PubMed: 12612996]
- 19.
- Nickeleit V, Steiger J, Mihatsch MJ. BK virus infection after kidney transplantation. Graft. 2002;5(DecemberSuppl):S46–S57.
- 20.
- Nickeleit V, Bubendorf L, Zeiler M. et al. Polyomavirus-infected decoy cells in urine cytology specimens of renal allograft recipients: Clinicopathological considerations. Lab Invest. 2002;82(1):82A.
- 21.
- Dalquen P, Kleiber B, Grilli B. et al. DNA image cytometry and fluorescence in situ hybridization for noninvasive detection of urothelial tumors in voided urine. Cancer. 2002;96(6):374–379. [PubMed: 12478686]
- 22.
- Bardales RH. Constituents of urinary tract specimens in the absence of diseaseIn: Bardales RH, ed.Practical Urologic CytopathologyNew York: Oxford University Press,200238–52.
- 23.
- Fogazzi GB, Cantu M, Saglimbeni L. ‘Decoy cells’ in the urine due to polyomavirus BK infection: Easily seen by phase-contrast microscopy. Nephrol Dial Transplant. 2001;16(7):1496–1498. [PubMed: 11427650]
- 24.
- Drachenberg RC, Drachenberg CB, Papadimitriou JC. et al. Morphological spectrum of polyoma virus disease in renal allografts: Diagnostic accuracy of urine cytology. Am J Transplant. 2001;1(4):373–381. [PubMed: 12099383]
- 25.
- Randhawa PS, Finkelstein S, Scantlebury V. et al. Human polyoma virus-associated interstitial nephritis in the allograft kidney. Transplantation. 1999;67(1):103–109. [PubMed: 9921805]
- 26.
- Drachenberg CB, Papadimitriou JC, Wali R. et al. Improved outcome of polyoma virus allograft nephropathy with early biopsy. Transplant Proc. 2004;36(3):758–759. [PubMed: 15110653]
- 27.
- Buehrig CK, Lager DJ, Stegall MD. et al. Influence of surveillance renal allograft biopsy on diagnosis and prognosis of polyomavirus-associated nephropathy. Kidney Int. 2003;64(2):665–673. [PubMed: 12846764]
- 28.
- Mayr M, Nickeleit V, Hirsch HH. et al. Polyomavirus BK nephropathy in a kidney transplant recipient: Critical issues of diagnosis and management. Am J Kidney Dis. 2001;38(3):E13. [PubMed: 11532715]
- 29.
- Nickeleit V, Klimkait T, Binet IF. et al. Testing for polyomavirus type BK DNA in plasma to identify renal-allograft recipients with viral nephropathy. N Engl J Med. 2000;342(18):1309–1315. [PubMed: 10793163]
- 30.
- Nickeleit V, Steiger J, Mihatsch MJ. Re: Noninvasive diagnosis of BK virus nephritis by measurement of messenger RNA for BK virus VP1. Transplantation. 2003;75(12):2160–2161. [PubMed: 12829935]
- 31.
- Hirsch HH, Knowles W, Dickenmann M. et al. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N Engl J Med. 2002;347(7):488–496. [PubMed: 12181403]
- 32.
- Randhawa P, Ho A, Shapiro R. et al. Correlates of quantitative measurement of BK polyomavirus (BKV) DNA with clinical course of BKV infection in renal transplant patients. J Clin Microbiol. 2004;42(3):1176–1180. [PMC free article: PMC356850] [PubMed: 15004071]
- 33.
- Limaye AP, Jerome KR, Kuhr CS. et al. Quantitation of BK virus load in serum for the diagnosis of BK virus-associated nephropathy in renal transplant recipients. J Infect Dis. 2001;183(11):1669–1672. [PubMed: 11343217]
- 34.
- Howell DN, Smith SR, Butterly DW. et al. Diagnosis and management of BK polyomavirus interstitial nephritis in renal transplant recipients. Transplantation. 1999;68(9):1279–1288. [PubMed: 10573064]
- 35.
- Ding R, Medeiros M, Dadhania D. et al. Noninvasive diagnosis of BK virus nephritis by measurement of messenger RNA for BK virus VP1 in urine. Transplantation. 2002;74(7):987–994. [PubMed: 12394843]
- 36.
- Biel SS, Nitsche A, Kurth A. et al. Detection of human polyomaviruses in urine from bone marrow transplant patients: Comparison of electron microscopy with PCR. Clin Chem. 2004;50(2):306–312. [PubMed: 14684621]
- Urine Cytology Findings of Polyomavirus Infections - Madame Curie Bioscience Dat...Urine Cytology Findings of Polyomavirus Infections - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...