NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Intrinsically curved DNA structures often occur in or around origins of DNA replication, regions that regulate transcription, and DNA recombination loci, and are found in a wide variety of cellular and viral genomes from bacteria to man. In bacterial promoters, bent DNA structures are often located from immediately upstream of the -35 hexamer to around position -100 relative to the transcription start site (+1). They have a range of functions: facilitating RNA polymerase binding to the promoter, transition from closed to open promoter complexes, or transcription factor binding. To perform these functions, in some cases intrinsically curved structures function together with DNA bends that are induced by binding of RNA polymerase, transcription factors, or nucleoid-associated proteins. This Chapter will describe how curved DNA structures are implicated in prokaryotic transcription.
Introduction
DNA can become bent either by an exterior force such as a protein binding, or by the nucleotide sequence per se. The former is called protein-induced DNA bending or simply DNA bending, and the latter is called bent DNA, curved DNA, or intrinsic DNA curvature. Initially, the formation of a stable bent configuration of DNA was proposed by Crick and Klug to explain the mechanism underlying DNA packaging into nucleosomes, which dates back to 1975.1 Several different models soon followed.2-6 Among them, the “wedge model” 5 and the “junction model”3 are famous as predicting intrinsic DNA curvatures6 (Chapter 2). The first naturally occurring curved DNA was discovered in 1982, by electrophoretic analyses of the minicircle fragments of mitochondrial (kinetoplast) DNA (k-DNA) from a parasite, Leishmania tarentolae.7 A DNA fragment from the organism migrated unusually slowly in non-denaturing polyacrylamide gels. This fragment contained a unique nucleotide sequence, with regularly distributed runs of adenines or thymines, with a periodicity of one run per helical repeat. Although such regular runs were soon proved to be the cause of intrinsic DNA curvature, the molecular mechanism to form curved DNA structure is still a matter of controversy.6,8-15
Since the initial discovery, curved DNA structures have been identified in a wide variety of cellular and viral genomes from bacteria to man. Interestingly, curved DNA structures often occur in or around origins of DNA replication,16-21 promoters and enhancers (Table 1; for eukaryotes see Table 1 in Chapter 5) and DNA recombination loci,22,23 irrespective of the origin of the DNA, suggesting that DNA curvature is important in many basic genetic processes (for reviews, see refs. 24-27). In order to reveal its role, a great many studies have been carried out. Here, focusing on the roles in prokaryotic transcription, we describe the fruits of these studies.
Table 1
Curved DNA structures found in or around prokaryotic promoters.
The Shape of Curved DNA
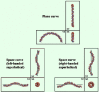
Naturally occurring curved DNA can adopt various conformations, which are determined by runs of adenines or thymines (A-tracts or T-tracts) as described above. When the A- or T-tracts occur with a periodicity almost equal to that of the DNA helical repeat length of around 10.5 bp, DNA forms a planar curve (i.e., a two-dimensional curve). However, if the tract periodicity is other than this, then either a right- or a left-handed superhelical conformation is formed.12 These three-dimensional (3D) structures are sometimes called "space curves" (fig. 1). When the periodicity is less than 10.5 bp (e.g., 9 or 10 bp), DNA adopts a left-handed superhelical conformation, and when it is more than 10.5 bp (e.g., 11 or 12 bp), DNA adopts a right-handed superhelical conformation. As the tract periodicity departs further from 10.5 bp, the helical axis becomes almost straight; e.g., periodicities of 5 to 7 bp result in a nearly straight (actually, a zigzag) trajectory of the helical axis.
Mapping Curved DNA Loci
Bacterial core promoters have conserved sequences at positions -35 (TTGACA) and -10 (TATAAT) relative to the transcription start site, which are recognized by RNA polymerase (RNAP) holoenzyme carrying a principal sigma factor such as sigma 70 (σ70) (Escherichia coli) or SigA (σA) (Bacillus subtilis). This section describes where curved DNAs occur, and a representative technique for studying DNA curvature.
Circular Permutation Analysis
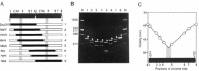
Circular permutation analysis, cyclization analysis, and electron microscopy are often used to find the position or magnitude of bends in DNA.6,28,29 Circular permutation analysis, devised by Wu and Crothers in 1984, has now become routine. It can reveal the position, magnitude, and direction of a DNA bend compared to known standards. Here, we will describe only this assay (see fig. 2). A DNA bend slows migration through a polyacrylamide gel. This phenomenon is more pronounced when the bend is in the center of a DNA fragment as opposed to near the ends. Thus to find the center of a bend, the DNA fragment of interest is first cloned as a tandem dimer, and is then cut with a series of restriction enzymes, each cutting once only within each fragment. The mobility (or relative mobility) of the fragments is plotted against the position of cutting. In the fragment which migrates most rapidly in the gel, the DNA cut was presumably closest to the center of the DNA bend.
Core Promoter Region
There are several reports of curved DNA in this region (Table 1). The E. coli lac core P1 promoter contains a slightly curved DNA around -31.30 In addition, promoters of E. coli glpF (a gene involved in facilitated diffusion of glycerol), helD (helicase IV), and proV (the first gene of the proU operon, involved in high-affinity transport of glycine, betaine and proline) are involved in curved DNA.31 The P2 promoter of the B. subtilis spo0F gene, which encodes a protein required to initiate sporulation, contains curved DNA centered around -10.32 The curved region contains the recognition site of phospho-Spo0A, a transcriptional regulator essential for the initiation of sporulation.
In the following promoters, absence of the consensus hexamer sequences may be offset by the presence of curved DNA. In E. coli, expression of a number of genes during the stationary phase is controlled by σS (KatF). Interestingly, the σS-regulated promoters do not present good consensus sequences in -10 and -35 regions, but are probably located in intrinsically curved DNA.33 Recently, a similar report has been presented on the Campylobacter jejuni promoters which are recognized by house-keeping sigma-factor, RpoD.34 C. jejuni is a human gastrointestinal pathogen. Petersen et al showed that the promoters recognized by RpoD do not contain the conserved -35 motif, but instead show very strong periodic variation in AT-content, and also semi-conserved T-tracts with a periodicity of 10-11 nucleotides that could form curved DNA. In cattle, Pasteurella haemolytica causes necrosis of the lung and the production of a fibrinocellular exudate. In the P. haemolytica leukotoxin promoter, the conserved -35 motif is also absent, and instead four repeats of CA6(C/T)A are present with approximately 10 bp periodicity. The repeats form a curved DNA structure, which influences transcription.35
Upstream of the Core Promoter
Based on computer modeling, an early study suggested that strong E. coli promoters harbor a bent DNA structure in their upstream sequences.36 Indeed, most of the curved DNA structures that have been experimentally identified are upstream of core promoters. Roughly, they fall into two groups: one located immediately upstream of the -35 hexamer, and the other further upstream. The first group includes promoters of the following genes: E. coli argU (arginine tRNA-4),31 the E. coli leuV tRNA operon,37 a Salmonella hisR tRNA operon,38 the E. coli rrnB rRNA operon,39 the E. coli rrnD rRNA operon,31 bla (β-lactamase gene) in E. coli plasmid pUC19,40,41 the Alu156 of Bacillus subtilis phage SP82,42 Clostridium perfringens plc [encoding phospholipase C (α-toxin)],43 and Microcystis aeruginosa (cyanobacterium) K-81 rpoD1 (encoding principal sigma factor, σA1).44
The second group includes DNA curvatures of E. coli argT, and the ilvlH operon (encodes acetohydroxyacid synthase III), with centers of curvature between -90 and -95,45 and at -120,46 respectively. M. aeruginosa K-81, a photosynthetic freshwater cyanobacterium, can grow between 5 and 38°C, with optimal growth at 28 to 30°C. Its psbA2 gene (encoding a core protein in photosystem II) contains a unique curved DNA upstream of the light-responsive promoter, from -180 to -140. Interestingly, the center of curvature depends on temperature: it was at bases -180, -160, and -140, at 50, 30, and 4°C, respectively.47 In many other organisms, the light-responsive genes also have curved DNA in the upstream region, e.g., in Synechocystis PCC6803 (blue-green algae), Cyanidium caldarium (red algae), Oryza sativa (plant, rice) and Nicotiana tabacum (plant, tobacco).48 This may suggest that the curved DNA is highly conserved among light responsive genes from cyanobacteria to higher plants. Streptococcus equisimilis H46A, a human serogroup C strain, is a potent producer of the plasminogen activator streptokinase. The streptokinase gene (skc) has an intrinsic DNA curvature located at -100.49
Some promoters have DNA curvatures in both regions. For example, the PctII promoter of plasmid pLS1, which can replicate in both Gram-positive and Gram-negative bacteria, carries two curvatures. One is in the proximal region (∼-50 to -80), and the other is in the distal region (∼-140 to -220).50
Transcription factor binding sites are often located in curved DNA, irrespective of the location of the curvature. Both E. coli lac and gal operons possess dual promoters, P1 and P2. In the lac operon, slight curves form around positions -30 and -100 relative to P1's transcription start point.30 The catabolite gene activator protein (CAP; sometimes known as CRP, the cyclic AMP receptor protein), a positive regulator responding to carbon source limitation,51 binds the region within this curvature.30,51 In the gal operon, the curved structure is located between -60 and -90 relative to P1's transcription start point. This region is slightly upstream of the CAP binding site, which is centered at -41.5.52-54 The E. coli ompF gene, coding for a major outer membrane protein, also carries curved DNA. The sequence causing the curve lies between -101 and -71, where two sets of periodically spaced A4 tracts are present. The binding site of OmpR (an activator) overlaps the curved DNA.55 In the E. coli rrnB P1 promoter, the curved DNA center overlaps one of three FIS (factor for inversion stimulation) binding sites.56,57 The B. subtilis phage Φ29 has a curved DNA that falls into this category.58 The promoters PA2b (the promoter of early genes) and PA3 (the promoter of late genes) partially overlap, and drive transcription in opposite directions. Two p4 binding sites, separated by 15 bp, are within a segment of curved DNA. When p4 binds, it increases the curvature.58 In the E. coli glnAp2 promoter (from the glnALG operon, encoding glutamine synthetase), computer simulation suggested that a 70 degree curve forms between the binding sites for the activator (nitrogen regulator I, NRI, = NtrC) and for σN (σ54)-RNAP.59 Similarly, in the Klebsiella pneumoniae nifLA promoter, a curved structure forms between two NtrC binding sites and the core promoter.60
Downstream of the Core Promoter
In this region, curved DNA is rare. One example is the slight curve in the operator region of the E. coli caa gene (colicin A).61,62 However, in this region, protein-induced DNA bends may be more important than intrinsic curvature, as regulators of transcription: when LexA (the repressor of the SOS system) binds here, it bends the DNA substantially.61,62
Role of the Curved DNA
First, we briefly describe how researchers have tried to understand the function of DNA conformation. We then detail how curved DNA is implicated in the transcription cascade.
Short History
The first functional analysis of DNA curvature was performed in 1984 using a promoter of a tRNA operon of Salmonella.38 A 3 bp deletion at position -70 disrupted a curved structure, and reduced in vivo transcription to 40%. This study suggested that curved DNA could control transcription. The E. coli argT promoter requires an upstream region for high in vivo activity. Deletion mutants were used to study this promoter,45 which required an upstream curved DNA region for high activity. Techniques using synthetic bent DNA are sometimes very useful, and have suggested that a DNA curvature close to the -35 hexamer is important.63 Furthermore, linker scanning mutations were used to study the DNA curvature just upstream of the -35 sequence of E. coli rrnB operon. They revealed that the angular orientation of the DNA curvature determines promoter activation.39
Insertion of a short DNA segment(s) into sites in or around the curved DNA region is also useful. In 1989, McAllister and Achberger investigated the function of curved DNA upstream of the Alu 156 promoter of B. subtilis phage SP82.42 By introducing short DNA fragments (6 to 29 bp) between the core promoter and the curved DNA, they changed the rotational phase between them. These changes correlated with the changes in promoter function in vivo. The most efficient mutant promoters contained insertions of 11 and 21 base pairs, and the least efficient promoters contained insertions of 15 and 25 base pairs. In vitro these mutations influenced the efficiency of RNA polymerase binding to the promoter. These findings demonstrate that the rotational phase between core promoter and the curved DNA is significant. The same methodology was used to study the promoter of C. perfringens plc gene and it was shown that upstream curved DNA stimulates transcription both in vivo and in vitro.43
In 1994, using deletion mutants, Pérez-Martín and Espinosa showed that curved DNA increases transcription from the PctII promoter of pLS1 in vivo and in vitro, apparently independently of any activator protein.50 Furthermore, an upstream curved DNA was replaced by the target sequence of IHF (integration host factor) or that of CopG (both are DNA-bending proteins), which activated transcription in the presence of these proteins but did not in their absence or deficiency. This study indicated that the curved conformation of DNA increases the number of contacts between the RNA polymerase and the promoter DNA, and that this increase is important in transcription initiation.
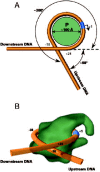
In order to alter shapes of DNA curvatures per se with minimal changes to sequence, short DNA segments were inserted into the center region of curved DNA in the pUC19 β-lactamase gene promoter,40 and into the cyanobacterium M. aeruginosa rpoD1 gene promoter.44 The resulting promoters were less active than the wild-type promoters, indicating that activity depended on the gross geometry. These promoters have right-handed curved DNA. Such DNAs are often located just upstream of promoters,64 but promoter DNA itself wraps around RNAP left-handedly (fig. 3).65-67 Thus it is tempting to speculate that if RNAP changed the writhe of the helical axis from right-handed to left-handed, it might deform the DNA and lead to local unwinding (and formation of an open promoter complex).64,65 To test this hypothesis, right-handed curved DNA, left-handed curved DNA, two-dimensionally curved DNA and straight DNA segments were synthesized and tested for their effect on transcription in vivo. Right-handed curved DNAs clearly facilitated formation of the open promoter complex and activated transcription.41 Curved DNA can also change its shape depending on temperature.68 Thus, temperature can be used to study the relationship between DNA conformation and function.69,70 Recently, a mechanism of transcriptional regulation has been proposed that depends on a temperature-induced conformational change (Chapter 4).
Mechanistic Roles
Transcription initiation involves several steps.71,72 Briefly, an RNAP binds to promoter (P), to yield RNAP-promoter closed complex (RPC) with an equilibrium constant KB. Next, RNAP melts approximately 14 bp of promoter DNA surrounding the transcription start site, with a rate constant k2, to yield an RNAP-promoter open complex (RPO). Subsequently, RNAP begins to synthesize RNA as an RNAP-promoter initial transcribing complex (RPITC). There are several abortive cycles of RNA synthesis, which yield RNA of 2∼8 nucleotides long, with a rate constant ki. When by chance a 9 nucleotide strand is synthesized, RNAP releases the promoter DNA (promoter clearance) and synthesizes RNA as an RNAP-DNA elongation complex (RDE). Bent DNA is implicated in the steps described below.
RNAP Binding
Several studies have indicated that curved DNA is important for binding RNAP to the promoter.70,73-75 In the gal P1 promoter, a point mutation from G•C to T•A at position -19, which abolishes P2 activity, enhances contact between the E. coli RNAP and the DNA between -49 and -54, and activates transcription from the P1 promoter, even in the absence of cAMP-CAP.73 This mutation generates a run of six consecutive thymines (5'-TTTTTT-3' is formed from 5'-TTTGTT-3'), which presumably influences the helical trajectory of the promoter, and helps RNAP binding. Hybrid E. coli λ phage promoters (λpR) have been created, carrying curved DNA from the B. subtilis phage SP82 promoter, Alu156, or Bal129, immediately upstream of the -35 region. These promoters bound E. coli RNAP more efficiently than did the original promoter. Interestingly, the upstream curved DNA wrapped around the RNAP in a nucleosomal-DNA-like fashion.75
The interaction between the α-subunit of RNAP and the curved DNA next to the core promoter may be important for initiating transcription. Many genes have an AT-rich upstream (UP) element upstream of the -35 hexamer, and this was originally identified in the E. coli rrnB P1 promoter. This element stimulates transcription through contact with the C-terminal domain of the α-subunit (α-CTD) of RNAP.76-79 The consensus sequence of the UP element is N2A3(A/T)(A/T)T(A/T)T4N2A4N3.80 This region behaved slightly abnormally during electrophoresis, suggesting that it may be curved. The curved conformation may influence interactions between the UP element and α-CTD.57 A study using the E. coli lacUV5 promoter revealed that in the RNAP-promoter open complex, α-CTD makes alternative nonspecific interactions with the DNA minor grooves at positions -43, -53, -63, -73, -83, and -93.81 Thus around the UP element, the conformation of DNA may assist the DNA to interact with α-CTD efficiently, and as a result, may influence transcription. However, it is difficult to distinguish the effects due to the shape of DNA from effects due to an AT-rich DNA sequence per se.78,82 None of the above studies paid attention to the 3D architecture of DNA. Interestingly, an in vitro experiment indicated that in the step of binding, E. coli RNAP favors DNA with a right-handed superhelical writhe.41
Formation of the Open Promoter Complex
After the RNAP holoenzyme binds to the promoter, approximately 14 bp of the promoter (between positions -11 and +3) melts.72 An early study hypothesized that DNA curvature allows upstream DNA to contact the promoter-bound RNAP, and that these interactions help the DNA for formation of the open promoter complex.42 Indeed, in some cases, curved DNA enhances formation of the open complex.41,54,83 For example, an in vitro experiment indicated that in the E. coli gal P1 promoter, the upstream curved DNA activates transcription by enhancing the rate of isomerization from the closed complex to the open complex (k2) at P1, both in the absence of the cAMP-CAP and in its presence (the study used mutants where the P2 promoter was silenced).54
DNA architectures are not necessarily optimally oriented towards the surface of the RNAP. The RNAP must therefore distort the DNA.84 Actually, the promoter DNA becomes bent when RNAP binds to it, and it wraps around the enzyme with a left-handed superhelical conformation (fig. 3).65-67,85,86 Energy invested in DNA bending could open the double helix.87 These considerations suggest that DNA curvatures can enhance stress, depending on the direction of the curvature, which explains well why many bacterial promoters have a right-handed curved DNA immediately upstream of the -35 region.64 This right-handed curvature presumably increases stress when the DNA wraps around RNAP, reversing its superhelical handedness. Indeed, when a synthetic right-handed curved DNA segment was placed immediately upstream of the -35 region of pUC19 β-lactamase promoter, it facilitated DNA melting at positions -11, -9, and +3 and activated transcription, compared to left-handed curved, two-dimensionally curved, and zigzag (straight) DNA segments.41 Positions -11 and +3 correspond to the limits of RNAP-induced promoter melting.72 Based on this putative mechanism, modulating the shape of DNA can also produce negative effects. A T5 tract introduced just upstream of the -35 hexamer reduced promoter activity.88 As another example, curved DNA inhibited transcription from the B. subtilis PA2b promoter, by reducing the ability of RNAP to form transcriptionally active open complexes (in this case, binding of RNAP to the promoter was also impaired).89 However, it is still not clear why DNA curvatures which are preset to modulate promoter melting, are usually located upstream of the core promoter.
Promoter Clearance
In the last stage of transcription initiation, promoter escape or promoter clearance occurs. “Escape” deals directly with issues affecting the downstream movement of a polymerase molecule. On the other hand, “clearance” implies that the polymerase moves far enough downstream to make the core promoter available to a second polymerase.90,91 These phenomena are also rate-limiting steps in transcription initiation. In the B. subtilis page Φ29 A2c promoter, preventing RNAP escape represses transcription.92 Are DNA curvatures involved in promoter escape or promoter clearance? The following study suggests they are. In a study using two promoters isolated from the B. subtilis bacteriophage SP82, curved DNA with a strong affinity for RNAP reduced transcription, while curved DNA with a weaker affinity stimulated transcription. 74 DNA curvatures may collaborate with H-NS to reduce promoter clearance - it is known that they can repress transcription (Chapter 4). Images from scanning force microscopy (SFM) showed that in an E. coli rrnB P1 promoter with an upstream DNA curvature, H-NS trapped RNA polymerase in the open initiation complex. The SFM images suggested that H-NS-mediated trapping of RNAP could prevent promoter clearance.93 To the best of our knowledge, a positive influence of DNA curvature has not been reported.
Transcription Factor Binding
If a curved DNA overlaps with some cis-DNA element, the curve's role may be to recruit trans-acting factors. In the M. aeruginosa psbA promoter, the curved region (approximately -180 to -140) is bound by a protein factor, and mutants having altered curvature had decreased basal transcription.48 E. coli OmpR and B. subtilis Spo0A are response regulators in bacterial two-component regulatory systems.94 Although OmpR can stimulate transcription of ompF (at low osmotic strength), and of ompC (at high osmotic strength), the OmpR binding site in the ompC promoter seems not to form a curved DNA. Thus, the presence or absence of curved DNA may be distinguished by OmpR in the differential activation of ompF and ompC promoters.55 The B. subtilis spo0F gene, which decides the cell fate in sporulation initiation, has a tandem promoter, P1 and P2. P1 is recognized by σA-RNAP during exponential cell growth, and P2 is recognized by σH-RNAP during initiation of sporulation. For Spo0A binding at P1, the center of DNA curvature is close to the 0A-boxes. Increased Spo0A binding to the 0A-boxes represses transcription from the upstream P1, and simultaneously induces transcription from the downstream P2.32 Curved DNA may facilitate Spo0A binding. However, no report has shown conclusively that curved DNA conformation per se is involved in factor binding.
Curved DNA as a Framework for Interaction Between Activator and RNAP
Intrinsically curved DNA is sometimes located between activator binding sites and promoters. Several examples indicate that these structures enable activator and RNAP to interact. In the σN-dependent glnAp2 or glnHp2 promoter of E. coli, the DNA between the NRI (NtrC)-binding site and the σN-recognition region must be either intrinsically curved, or curved by binding of the integration host factor, IHF. The glnAp2 has an intrinsic DNA curvature in the relevant region. Although glnHp2 does not carry curved DNA, it has an IHF-binding site in the relevant region. The DNA bend allows the activator to contact the σN-RNAP-promoter complex. This activates transcription by catalyzing the isomerization of the closed σN-RNAP-promoter complex to form an open complex.59,95,96
However, when supercoiled DNA templates are used, transcription can be initiated even in the absence of such a bend,96 meaning that the spatial alignment between NRI and RNAP on the templates may be significant. Similarly, in the K. pneumoniae nifLA promoter, an activator protein NtrC may interact with the σN-RNAP holoenzyme bound to the promoter with the help of an intervening curved DNA.60 As another example, in the E. coli rrnB P1 promoter, the rotational phase between the FIS binding site and the promoter is important in activating transcription.97 The curvature between these regions may also help FIS and RNAP to spatially position themselves correctly.
Other Roles
Curved DNA can also play a role in packaging of genomes. In eukaryotes, this role is played in nucleosome or chromatin formation (see Chapter 13). In E. coli, some nucleoid-associated proteins, such as H-NS (histone-like nucleoid-structuring protein), CbpA (curved DNA-binding protein A), Hfq (host factor for phage Qβ), and IciA (inhibitor of chromosome initiation A) preferentially bind to curved DNA.98-100 Of these, H-NS can condense DNA both in vitro and in vivo101,102 and regulates expression of a number of genes,26,103 which are described in Chapter 4.
Curved DNA may also modulate the geometry of promoters in collaboration with structure-specific transcriptional regulators. The B. subtilis LrpC protein, which belongs to the LrpC/AsnC family of transcriptional regulators, forms stable complexes with curved DNAs. Interestingly, LrpC proteins wrap DNA to form nucleosome-like structures (but containing positively supercoiled DNA), and it is speculated that these could regulate transcription.104 As another function, temperature-dependent conformational changes of bent DNA, as observed for M. aeruginosa psbA2 gene,47 can regulate transcription of several genes (Chapter 4).
Intrinsic DNA Curvature and Transcription-Factor Induced DNA Bend
A great many DNA binding proteins, including transcription factors, from both eukaryotes and prokaryotes, bend DNA on binding.26,105-107 E. coli CAP, FIS and IHF, and B. subtilis Φ29 p4 protein are well known to induce DNA bends (for a review, see ref. 26). CAP activates transcription through protein-protein interaction with RNAP.108,109 The CAP-induced DNA bend might be required to create the appropriate geometry for this interaction.110 IHF and FIS are nucleoid-associated proteins. IHF is distributed uniformly within the nucleoid, while FIS accumulates at specific loci.111 They help formation of DNA micro-loops and/or nucleoprotein complexes.112-114 RNAP itself also induces DNA bends.115,116 In addition, there are many reports of repressors causing DNA to bend.116-121
For transcriptional activation, it is thought that induced DNA bends promote polymerase binding, protein-protein interactions, wrapping of DNA around the RNAP or multi-component DNA-protein complexes, local unwinding of DNA, and in some cases dissociation of nucleoprotein complexes.106,122 Intrinsic DNA bends can also generate most of these effects. Indeed, CAP-induced DNA bends were functionally substituted by replacing the CAP binding site of the gal promoter or the lac promoter with synthetic bent DNA sequences.83,123 Also, the IHF binding site was functionally replaced by intrinsically bent DNA sequences.124 Conversely, as described above, intrinsic DNA curvature can be functionally replaced by protein-induced DNA bends.50 These studies shed light on the significance of DNA bend per se in transcription initiation. The same is true for transcriptional repression. The p4 bends its target site on binding, excluding B. subtilis σA-RNA polymerase from the PA2b promoter, and directing it to the PA3 promoter. As a result, transcription from the Φ29 PA2b promoter is repressed. Interestingly, the p4-induced DNA bend can be functionally substituted by intrinsically bent DNA.89
As described above, intrinsic DNA bends and protein-induced DNA bends can play the same role. In some cases, they collaborate. In E. coli, the ilvPG2 promoter drives transcription of the ilvGMEDA operon whose products are required for the biosynthesis of L-leucine, L-isoleucine and L-valine. The promoter is activated both by intrinsic bend around position -50, and by IHF-induced DNA bend around -90.125 The DNA geometry and localized destabilization of the DNA helix, generated by these two bends, might facilitate RNAP-promoter interaction and DNA unwinding. Proteins that bend intrinsically curved DNA structures toward different direction can also facilitate DNA unwinding as discussed for open promoter complex formation. This phenomenon is another example of the collaboration. The general importance of bending induced by transcription factors is described in Chapter 11.
Conclusion
Intrinsically curved DNA structures can modulate transcription initiation. Although such structures have various functions in the transcription initiation cascade, it appears that they mainly work by helping RNAP to bind to the promoter, and by facilitating formation of the open promoter complex. Bacteria are able to control promoter activity by carefully tuning the three-dimensional architecture of the promoter.
Acknowledgements
The authors would like to acknowledge the contribution of Yoshiro Fukue. Our own studies reported in this chapter were partly supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
References
- 1.
- Crick FH, Klug A. Kinky helix. Nature. 1975;255:530–533. [PubMed: 1095931]
- 2.
- Sobell HM, Tsai CC, Gilbert SG. et al. Organization of DNA in chromatin. Proc Natl Acad Sci U S A. 1976;73:3068–3072. [PMC free article: PMC430931] [PubMed: 1067602]
- 3.
- Selsing E, Wells RD, Alden CJ. et al. Bent DNA: visualization of a base-paired and stacked A-B conformational junction. J Biol Chem. 1979;254:5417–5422. [PubMed: 447660]
- 4.
- Zhurkin VB, Lysov YP, Ivanov VI. Anisotropic flexibility of DNA and the nucleosomal structure. Nucleic Acids Res. 1979;6:1081–1096. [PMC free article: PMC327755] [PubMed: 440969]
- 5.
- Trifonov EN, Sussman JL. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci U S A. 1980;77:3816–3820. [PMC free article: PMC349717] [PubMed: 6933438]
- 6.
- Wu HM, Crothers DM. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984;308:509–513. [PubMed: 6323997]
- 7.
- Marini JC, Levene SD, Crothers DM. et al. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982;79:7664–7668. [PMC free article: PMC347408] [PubMed: 16593261]
- 8.
- Hagerman PJ. Sequence-directed curvature of DNA. Nature. 1986;321:449–450. [PubMed: 3713816]
- 9.
- Diekmann S. Sequence specificity of curved DNA. FEBS Lett. 1986;195:53–56. [PubMed: 3943623]
- 10.
- Ulanovsky LE, Trifonov EN. Estimation of wedge components in curved DNA. Nature. 1987;326:720–722. [PubMed: 3561514]
- 11.
- Koo HS, Crothers DM. Calibration of DNA curvature and a unified description of sequence-directed bending. Proc Natl Acad Sci U S A. 1988;85:1763–1767. [PMC free article: PMC279859] [PubMed: 3162306]
- 12.
- Calladine CR, Drew HR, McCall MJ. The intrinsic curvature of DNA in solution. J Mol Biol. 1988;201:127–137. [PubMed: 3418695]
- 13.
- Olson WK, Marky NL, Jernigan RL. et al. Influence of fluctuations on DNA curvature. A comparison of flexible and static wedge models of intrinsically bent DNA. J Mol Biol. 1993;232:530–554. [PubMed: 8345522]
- 14.
- Mack DR, Chiu TK, Dickerson RE. Intrinsic bending and deformability at the T-A step of CCTTTAAAGG: a comparative analysis of T-A and A-T steps within A-tracts. J Mol Biol. 2001;312:1037–1049. [PubMed: 11580248]
- 15.
- Barbic A, Zimmer DP, Crothers DM. Structural origins of adenine-tract bending. Proc Natl Acad Sci U S A. 2003;100:2369–2373. [PMC free article: PMC151347] [PubMed: 12586860]
- 16.
- Zahn K, Blattner FR. Direct evidence for DNA bending at the λ replication origin. Science. 1987;236:416–422. [PubMed: 2951850]
- 17.
- Hertz GZ, Young MR, Mertz JE. The A+T-rich sequence of the simian virus 40 origin is essential for replication and is involved in bending of the viral DNA. J Virol. 1987;61:2322–2325. [PMC free article: PMC283701] [PubMed: 3035231]
- 18.
- Williams JS, Eckdahl TT, Anderson JN. Bent DNA functions as a replication enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2763–2769. [PMC free article: PMC363493] [PubMed: 3043195]
- 19.
- Du C, Sanzgiri RP, Shaiu WL. et al. Modular structural elements in the replication origin region of Tetrahymena rDNA. Nucleic Acids Res. 1995;23:1766–1774. [PMC free article: PMC306934] [PubMed: 7784181]
- 20.
- Polaczek P, Kwan K, Liberies DA. et al. Role of architectural elements in combinatorial regulation of initiation of DNA replication in Escherichia coli. Mol Microbiol. 1997;26:261–275. [PubMed: 9383152]
- 21.
- Kusakabe T, Sugimoto Y, Hirota Y. et al. Isolation of replicational cue elements from a library of bent DNAs of Aspergillus oryzae. Mol Biol Rep. 2000;27:13–19. [PubMed: 10939521]
- 22.
- Milot E, Belmaaza A, Wallenburg JC. et al. Chromosomal illegitimate recombination in mammalian cells is associated with intrinsically bent DNA elements. EMBO J. 1992;11:5063–5070. [PMC free article: PMC556984] [PubMed: 1464328]
- 23.
- Mazin A, Milot E, Devoret R. et al. KIN17, a mouse nuclear protein, binds to bent DNA fragments that are found at illegitimate recombination junctions in mammalian cells. Mol Gen Genet. 1994;244:435–438. [PubMed: 8078469]
- 24.
- Travers A. DNA structure. Curves with a function. Nature. 1989;341:184–185. [PubMed: 2779666]
- 25.
- Hagerman PJ. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. [PubMed: 2197990]
- 26.
- Pérez-Martín J, Rojo F, de LorenzoV. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol Rev. 1994;58:268–290. [PMC free article: PMC372964] [PubMed: 8078436]
- 27.
- Ohyama T. Intrinsic DNA bends: an organizer of local chromatin structure for transcription. Bioessays. 2001;23:708–715. [PubMed: 11494319]
- 28.
- Levene SD, Crothers DM. Ring closure probabilities for DNA fragments by Monte Carlo simulation. J Mol Biol. 1986;189:61–72. [PubMed: 3783680]
- 29.
- Laundon CH, Griffith JD. Curved helix segments can uniquely orient the topology of supertwisted DNA. Cell. 1988;52:545–549. [PubMed: 2830027]
- 30.
- Liu-Johnson HN, Gartenberg MR, Crothers DM. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986;47:995–1005. [PubMed: 3536129]
- 31.
- Tanaka K, Muramatsu S, Yamada H. et al. Systematic characterization of curved DNA segments randomly cloned from Escherichia coli and their functional significance. Mol Gen Genet. 1991;226:367–376. [PubMed: 1903834]
- 32.
- Asayama M, Yamamoto A, Kobayashi Y. Dimer form of phosphorylated Spo0A, a transcriptional regulator, stimulates the spo0F transcription at the initiation of sporulation in Bacillus subtilis. J Mol Biol. 1995;250:11–23. [PubMed: 7541470]
- 33.
- Espinosa-Urgel M, Tormo A. σS-dependent promoters in Escherichia coli are located in DNA regions with intrinsic curvature. Nucleic Acids Res. 1993;21:3667–3670. [PMC free article: PMC309863] [PubMed: 8367283]
- 34.
- Petersen L, Larsen TS, Ussery DW. et al. RpoD promoters in Campylobacter jejuni exhibit a strong periodic signal instead of a -35 box. J Mol Biol. 2003;326:1361–1372. [PubMed: 12595250]
- 35.
- Highlander SK, Weinstock GM. Static DNA bending and protein interactions within the Pasteurella haemolytica leukotoxin promoter region: development of an activation model for leukotoxin transcriptional control. DNA Cell Biol. 1994;13:171–181. [PubMed: 8179822]
- 36.
- Plaskon RR, Wartell RM. Sequence distributions associated with DNA curvature are found upstream of strong E. coli promoters. Nucleic Acids Res. 1987;15:785–796. [PMC free article: PMC340467] [PubMed: 3547329]
- 37.
- Bauer BF, Kar EG, Elford RM. et al. Sequence determinants for promoter strength in the leuV operon of Escherichia coli. Gene. 1988;63:123–134. [PubMed: 2454871]
- 38.
- Bossi L, Smith DM. Conformational change in the DNA associated with an unusual promoter mutation in a tRNA operon of Salmonella. Cell. 1984;39:643–652. [PubMed: 6096016]
- 39.
- Zacharias M, Theissen G, Bradaczek C. et al. Analysis of sequence elements important for the synthesis and control of ribosomal RNA in E. coli. Biochimie. 1991;73:699–712. [PubMed: 1764516]
- 40.
- Ohyama T, Nagumo M, Hirota Y. et al. Alteration of the curved helical structure located in the upstream region of the β-lactamase promoter of plasmid pUC19 and its effect on transcription. Nucleic Acids Res. 1992;20:1617–1622. [PMC free article: PMC312246] [PubMed: 1579452]
- 41.
- Hirota Y, Ohyama T. Adjacent upstream superhelical writhe influences an Escherichia coli promoter as measured by in vivo strength and in vitro open complex formation. J Mol Biol. 1995;254:566–578. [PubMed: 7500334]
- 42.
- McAllister CF, Achberger EC. Rotational orientation of upstream curved DNA affects promoter function in Bacillus subtilis. J Biol Chem. 1989;264:10451–10456. [PubMed: 2543669]
- 43.
- Matsushita C, Matsushita O, Katayama S. et al. An upstream activating sequence containing curved DNA involved in activation of the Clostridium perfringens plc promoter. Microbiology. 1996;142:2561–2566. [PubMed: 8828224]
- 44.
- Asayama M, Hayasaka Y, Kabasawa M, Shirai M, Ohyama A. An intrinsic DNA curvature found in the cyanobacterium Microcystis aeruginosa K-81 affects the promoter activity of rpoD1 encoding a principal σ factor. J Biochem (Tokyo). 1999;125:460–468. [PubMed: 10050033]
- 45.
- Hsu LM, Giannini JK, Leung TW. et al. Upstream sequence activation of Escherichia coli argT promoter in vivo and in vitro. Biochemistry. 1991;30:813–822. [PubMed: 1988068]
- 46.
- Wang Q, Albert FG, Fitzgerald DJ. et al. Sequence determinants of DNA bending in the ilvlH promoter and regulatory region of Escherichia coli. Nucleic Acids Res. 1994;22:5753–5760. [PMC free article: PMC310143] [PubMed: 7838732]
- 47.
- Agrawal GK, Asayama M, Shirai M. A novel bend of DNA CIT: changeable bending-center sites of an intrinsic curvature under temperature conditions. FEMS Microbiol Lett. 1997;147:139–145. [PubMed: 9037772]
- 48.
- Asayama M, Kato H, Shibato J. et al. The curved DNA structure in the 5'-upstream region of the light-responsive genes: its universality, binding factor and function for cyanobacterial psbA transcription. Nucleic Acids Res. 2002;30:4658–4666. [PMC free article: PMC140650] [PubMed: 12409456]
- 49.
- Groß S, Gase K, Malke H. Localization of the sequence-determined DNA bending center upstream of the streptokinase gene skc. Arch Microbiol. 1996;166:116–121. [PubMed: 8929127]
- 50.
- Pérez-Martín J, Espinosa M. Correlation between DNA bending and transcriptional activation at a plasmid promoter. J Mol Biol. 1994;241:7–17. [PubMed: 8051709]
- 51.
- Kolb A, Busby S, Buc H. et al. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. [PubMed: 8394684]
- 52.
- Taniguchi T, O'Neill M, de CrombruggheB. Interaction site of Escherichia coli cyclic AMP receptor protein on DNA of galactose operon promoters. Proc Natl Acad Sci U S A. 1979;76:5090–5094. [PMC free article: PMC413085] [PubMed: 228278]
- 53.
- Taniguchi T, de CrombruggheB. Interactions of RNA polymerase and the cyclic AMP receptor protein on DNA of the E. coli galactose operon. Nucleic Acids Res. 1983;11:5165–5180. [PMC free article: PMC326245] [PubMed: 6308575]
- 54.
- Lavigne M, Herbert M, Kolb A. et al. Upstream curved sequences influence the initiation of transcription at the Escherichia coli galactose operon. J Mol Biol. 1992;224:293–306. [PubMed: 1313883]
- 55.
- Mizuno T. Static bend of DNA helix at the activator recognition site of the ompF promoter in Escherichia coli. Gene. 1987;54:57–64. [PubMed: 3301541]
- 56.
- Ross W, Thompson JF, Newlands JT. et al. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–3742. [PMC free article: PMC552129] [PubMed: 2209559]
- 57.
- Gaal T, Rao L, Estrem ST. et al. Localization of the intrinsically bent DNA region upstream of the E. coli rrnB P1 promoter. Nucleic Acids Res. 1994;22:2344–2350. [PMC free article: PMC523693] [PubMed: 8036162]
- 58.
- Rojo F, Zaballos A, Salas M. Bend induced by the phage Φ29 transcriptional activator in the viral late promoter is required for activation. J Mol Biol. 1990;211:713–725. [PubMed: 2107318]
- 59.
- Carmona M, Magasanik B. Activation of transcription at σ54-dependent promoters on linear templates requires intrinsic or induced bending of the DNA. J Mol Biol. 1996;261:348–356. [PubMed: 8780778]
- 60.
- Cheema AK, Choudhury NR, Das HK. A- and T-tract-mediated intrinsic curvature in native DNA between the binding site of the upstream activator NtrC and the nifLA promoter of Klebsiella pneumoniae facilitates transcription. J Bacteriol. 1999;181:5296–5302. [PMC free article: PMC94035] [PubMed: 10464200]
- 61.
- Lloubès R, Granger-Schnarr M, Lazdunski C. et al. LexA repressor induces operator-dependent DNA bending. J Mol Biol. 1988;204:1049–1054. [PubMed: 3065515]
- 62.
- Lloubès R, Lazdunski C, Granger-Schnarr M. et al. DNA sequence determinants of LexA-induced DNA bending. Nucleic Acids Res. 1993;21:2363–2367. [PMC free article: PMC309533] [PubMed: 8506133]
- 63.
- Collis CM, Molloy PL, Both GW. et al. Influence of the sequence-dependent flexure of DNA on transcription in E. coli. Nucleic Acids Res. 1989;17:9447–9468. [PMC free article: PMC335145] [PubMed: 2685760]
- 64.
- Travers AA. Why bend DNA? Cell. 1990;60:177–180. [PubMed: 2404609]
- 65.
- Amouyal M, Buc H. Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J Mol Biol. 1987;195:795–808. [PubMed: 3309341]
- 66.
- Coulombe B, Burton ZF. DNA bending and wrapping around RNA polymerase: a “revolutionary” model describing transcriptional mechanisms. Microbiol Mol Biol Rev. 1999;63:457–478. [PMC free article: PMC98973] [PubMed: 10357858]
- 67.
- Rivetti C, Guthold M, Bustamante C. Wrapping of DNA around the E. coli RNA polymerase open promoter complex. EMBO J. 1999;18:4464–4475. [PMC free article: PMC1171521] [PubMed: 10449412]
- 68.
- Diekmann S. Temperature and salt dependence of the gel migration anomaly of curved DNA fragments. Nucleic Acids Res. 1987;15:247–265. [PMC free article: PMC340408] [PubMed: 3029673]
- 69.
- Ohyama T. Bent DNA in the human adenovirus type 2 E1A enhancer is an architectural element for transcription stimulation. J Biol Chem. 1996;271:27823–27828. [PubMed: 8910380]
- 70.
- Katayama S, Matsushita O, Jung CM. et al. Promoter upstream bent DNA activates the transcription of the Clostridium perfringens phospholipase C gene in a low temperature-dependent manner. EMBO J. 1999;18:3442–3450. [PMC free article: PMC1171423] [PubMed: 10369683]
- 71.
- Gussin GN. Kinetic analysis of RNA polymerase-promoter interactions. Methods Enzymol. 1996;273:45–59. [PubMed: 8791598]
- 72.
- Ebright RH. RNA polymerase-DNA interaction: structures of intermediate, open, and elongation complexes. Cold Spring Harb Symp Quant Biol. 1998;63:11–20. [PubMed: 10384266]
- 73.
- Busby S, Spassky A, Chan B. RNA polymerase makes important contacts upstream from base pair -49 at the Escherichia coli galactose operon P1 promoter. Gene. 1987;53:145–152. [PubMed: 3038692]
- 74.
- McAllister CF, Achberger EC. Effect of polyadenine-containing curved DNA on promoter utilization in Bacillus subtilis. J Biol Chem. 1988;263:11743–11749. [PubMed: 3136165]
- 75.
- Nickerson CA, Achberger EC. Role of curved DNA in binding of Escherichia coli RNA polymerase to promoters. J Bacteriol. 1995;177:5756–5761. [PMC free article: PMC177394] [PubMed: 7592319]
- 76.
- Ross W, Gosink KK, Salomon J. et al. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science. 1993;262:1407–1413. [PubMed: 8248780]
- 77.
- Estrem ST, Ross W, Gaal T. et al. Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase α subunit. Genes Dev. 1999;13:2134–2147. [PMC free article: PMC316962] [PubMed: 10465790]
- 78.
- Gourse RL, Ross W, Gaal T. UPs and downs in bacterial transcription initiation: the role of the α subunit of RNA polymerase in promoter recognition. Mol Microbiol. 2000;37:687–695. [PubMed: 10972792]
- 79.
- Ross W, Ernst A, Gourse RL. Fine structure of E. coli RNA polymerase-promoter interactions: α subunit binding to the UP element minor groove. Genes Dev. 2001;15:491–506. [PMC free article: PMC312649] [PubMed: 11238372]
- 80.
- Estrem ST, Gaal T, Ross W. et al. Identification of an UP element consensus sequence for bacterial promoters. Proc Natl Acad Sci U S A. 1998;95:9761–9766. [PMC free article: PMC21410] [PubMed: 9707549]
- 81.
- Naryshkin N, Revyakin A, Kim Y. et al. Structural organization of the RNA polymerase-promoter open complex. Cell. 2000;101:601–611. [PubMed: 10892647]
- 82.
- Aiyar SE, Gourse RL, Ross W. Upstream A-tracts increase bacterial promoter activity through interactions with the RNA polymerase α subunit. Proc Natl Acad Sci U S A. 1998;95:14652–14657. [PMC free article: PMC24504] [PubMed: 9843944]
- 83.
- Gartenberg MR, Crothers DM. Synthetic DNA bending sequences increase the rate of in vitro transcription initiation at the Escherichia coli lac promoter. J Mol Biol. 1991;219:217–230. [PubMed: 1645411]
- 84.
- Borowiec JA, Gralla JD. High-resolution analysis of lac transcription complexes inside cells. Biochemistry. 1986;25:5051–5057. [PubMed: 3533143]
- 85.
- Mekler V, Kortkhonjia E, Mukhopadhyay J. et al. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. [PubMed: 11893332]
- 86.
- Murakami KS, Masuda S, Campbell EA. et al. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. [PubMed: 12016307]
- 87.
- Ramstein J, Lavery R. Energetic coupling between DNA bending and base pair opening. Proc Natl Acad Sci U S A. 1988;85:7231–7235. [PMC free article: PMC282158] [PubMed: 3174629]
- 88.
- Lozinski T, Adrych-Rozek K, Markiewicz WT. et al. Effect of DNA bending in various regions of a consensus-like Escherichia coli promoter on its strength in vivo and structure of the open complex in vitro. Nucleic Acids Res. 1991;19:2947–2953. [PMC free article: PMC328256] [PubMed: 2057353]
- 89.
- Rojo F, Salas M. A DNA curvature can substitute phage Φ29 regulatory protein p4 when acting as a transcriptional repressor. EMBO J. 1991;10:3429–3438. [PMC free article: PMC453071] [PubMed: 1655421]
- 90.
- Gralla JD. Promoter recognition and mRNA initiation by Escherichia coli Eσ70. Methods Enzymol. 1990;185:37–54. [PubMed: 2199788]
- 91.
- Hsu LM. Promoter clearance and escape in prokaryotes. Biochim Biophys Acta. 2002;1577:191–207. [PubMed: 12213652]
- 92.
- Salas M. Control mechanisms of bacteriophage Φ29 DNA expression. Int Microbiol. 1998;1:307–310. [PubMed: 10943379]
- 93.
- Dame RT, Wyman C, Wurm R. et al. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J Biol Chem. 2002;277:2146–2150. [PubMed: 11714691]
- 94.
- Stock JB, Stock AM, Mottonen JM. Signal transduction in bacteria. Nature. 1990;344:395–400. [PubMed: 2157156]
- 95.
- Claverie-Martin F, Magasanik B. Positive and negative effects of DNA bending on activation of transcription from a distant site. J Mol Biol. 1992;227:996–1008. [PubMed: 1433305]
- 96.
- Carmona M, Claverie-Martin F, Magasanik B. DNA bending and the initiation of transcription at σ54-dependent bacterial promoters. Proc Natl Acad Sci USA. 1997;94:9568–9572. [PMC free article: PMC23220] [PubMed: 9275163]
- 97.
- Newlands JT, Josaitis CA, Ross W. et al. Both fis-dependent and factor-independent upstream activation of the rrnB P1 promoter are face of the helix dependent. Nucleic Acids Res. 1992;20:719–726. [PMC free article: PMC312010] [PubMed: 1542568]
- 98.
- Yamada H, Muramatsu S, Mizuno T. An Escherichia coli protein that preferentially binds to sharply curved DNA. J Biochem (Tokyo). 1990;108:420–425. [PubMed: 2126011]
- 99.
- Ueguchi C, Kakeda M, Yamada H. et al. An analogue of the DnaJ molecular chaperone in Escherichia coli. Proc Natl Acad Sci U S A. 1994;91:1054–1058. [PMC free article: PMC521452] [PubMed: 8302830]
- 100.
- Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J Biol Chem. 1999;274:33105–33113. [PubMed: 10551881]
- 101.
- Spassky A, Rimsky S, Garreau H. et al. H1a, an E. coli DNA-binding protein which accumulates in stationary phase, strongly compacts DNA in vitro. Nucleic Acids Res. 1984;12:5321–5340. [PMC free article: PMC318922] [PubMed: 6379600]
- 102.
- Spurio R, Durrenberger M, Falconi M. et al. Lethal overproduction of the Escherichia coli nucleoid protein H-NS: ultramicroscopic and molecular autopsy. Mol Gen Genet. 1992;231:201–211. [PubMed: 1310520]
- 103.
- Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. [PubMed: 9140961]
- 104.
- Beloin C, Jeusset J, Révet B. et al. Contribution of DNA conformation and topology in right-handed DNA wrapping by the Bacillus subtilis LrpC protein. J Biol Chem. 2003;278:5333–5342. [PubMed: 12458218]
- 105.
- Horikoshi M, Bertuccioli C, Takada R. et al. Transcription factor TFIID induces DNA bending upon binding to the TATA element. Proc Natl Acad Sci U S A. 1992;89:1060–1064. [PMC free article: PMC48385] [PubMed: 1736286]
- 106.
- van der Vliet PC, Verrijzer CP. Bending of DNA by transcription factors. Bioessays. 1993;15:25–32. [PubMed: 8466473]
- 107.
- Werner MH, Gronenborn AM, Clore GM. Intercalation, DNA kinking, and the control of transcription. Science. 1996;271:778–784. [PubMed: 8628992]
- 108.
- Zhou Y, Zhang X, Ebright RH. Identification of the activating region of catabolite gene activator protein (CAP): isolation and characterization of mutants of CAP specifically defective in transcription activation. Proc Natl Acad Sci U S A. 1993;90:6081–6085. [PMC free article: PMC46871] [PubMed: 8392187]
- 109.
- Heyduk T, Lee JC, Ebright YW. et al. CAP interacts with RNA polymerase in solution in the absence of promoter DNA. Nature. 1993;364:548–549. [PubMed: 8393148]
- 110.
- Kapanidis AN, Ebright YW, Ludescher RD. et al. Mean DNA bend angle and distribution of DNA bend angles in the CAP-DNA complex in solution. J Mol Biol. 2001;312:453–468. [PubMed: 11563909]
- 111.
- Azam TA, Hiraga S, Ishihama A. Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells. 2000;5:613–626. [PubMed: 10947847]
- 112.
- Goosen N, van de Putte P. The regulation of transcription initiation by integration host factor. Mol Microbiol. 1995;16:1–7. [PubMed: 7651128]
- 113.
- Travers A, Muskhelishvili G. DNA microloops and microdomains: a general mechanism for transcription activation by torsional transmission. J Mol Biol. 1998;279:1027–1043. [PubMed: 9642081]
- 114.
- Travers A, Schneider R, Muskhelishvili G. DNA supercoiling and transcription in Escherichia coli: The FIS connection. Biochimie. 2001;83:213–217. [PubMed: 11278071]
- 115.
- Kuhnke G, Fritz HJ, Ehring R. Unusual properties of promoter-up mutations in the Escherichia coli galactose operon and evidence suggesting RNA polymerase-induced DNA bending. EMBO J. 1987;6:507–513. [PMC free article: PMC553423] [PubMed: 3034593]
- 116.
- Kuhnke G, Theres C, Fritz HJ. et al. RNA polymerase and gal repressor bind simultaneously and with DNA bending to the control region of the Escherichia coli galactose operon. EMBO J. 1989;8:1247–1255. [PMC free article: PMC400941] [PubMed: 2663472]
- 117.
- Pérez-Martín J, del SolarGH, Lurz R. et al. Induced bending of plasmid pLS1 DNA by the plasmid-encoded protein RepA. J Biol Chem. 1989;264:21334–21339. [PubMed: 2592378]
- 118.
- Pérez-Martín J, Espinosa M. The RepA repressor can act as a transcriptional activator by inducing DNA bends. EMBO J. 1991;10:1375–1382. [PMC free article: PMC452798] [PubMed: 2026140]
- 119.
- Ansari AZ, Chael ML, O'Halloran TV. Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg-MerR. Nature. 1992;355:87–89. [PubMed: 1731201]
- 120.
- Wang L, Winans SC. High angle and ligand-induced low angle DNA bends incited by OccR lie in the same plane with OccR bound to the interior angle. J Mol Biol. 1995;253:32–38. [PubMed: 7473714]
- 121.
- Tian G, Lim D, Carey J. et al. Binding of the arginine repressor of Escherichia coli K-12 to its operator sites. J Mol Biol. 1992;226:387–397. [PubMed: 1640457]
- 122.
- Zinkel SS, Crothers DM. Catabolite activator protein-induced DNA bending in transcription initiation. J Mol Biol. 1991;219:201–215. [PubMed: 1645410]
- 123.
- Bracco L, Kotlarz D, Kolb A. et al. Synthetic curved DNA sequences can act as transcriptional activators in Escherichia coli. EMBO J. 1989;8:4289–4296. [PMC free article: PMC401636] [PubMed: 2512122]
- 124.
- Pérez-Martín J, Timmis KN, de Lorenzo V. Co-regulation by bent DNA. Functional substitutions of the integration host factor site at σ54-dependent promoter Pu of the upper-TOL operon by intrinsically curved sequences. J Biol Chem. 1994;269:22657–22662. [PubMed: 8077217]
- 125.
- Pagel JM, Winkelman JW, Adams CW. et al. DNA topology-mediated regulation of transcription initiation from the tandem promoters of the ilvGMEDA operon of Escherichia coli. J Mol Biol. 1992;224:919–935. [PubMed: 1569580]
- Curved DNA and Prokaryotic Promoters: A Mechanism for Activation of Transcriptio...Curved DNA and Prokaryotic Promoters: A Mechanism for Activation of Transcription - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...