NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
Storing lipids as a reservoir for energy or the anabolism of elementary metabolites is a common feature of life in organisms from bacteria to humans. The chemical probes yielded by this project should serve as useful tools for providing a better understanding of cellular and organismic lipid storage on a functional and evolutionary level. Furthermore, active substances might result in the identification of lead compounds for the treatment of emerging lipid storage-associated diseases, including atherosclerosis, diabetes, or obesity. Also, the present assay will establish a profile of compounds that modulate this ubiquitous area of biology. The probe described here came from initial screening of known bioactive compound collections within the MLSMR. By combining the small molecule screening results with lipid metabolism modulating gene functions identified in the genome-wide RNAi screen, we were able to identify and link the Golgi Type I coat proteins known as cotamer, COPI to lipid storage. The developed probe, ML084 (CID-310557; Exo1), was shown to induce a lipid storage phenotype in cells through modulation of the protein trafficking pathways.
Assigned Assay Grant #: 1 R03 MH085686-01
Screening Center Name & PI: NIH Chemical Genomics Center, Christopher Austin
Chemistry Center Name & PI: NIH Chemical Genomics Center, Christopher Austin
Assay Submitter & Institution: Dr. Mathias Beller, Max Planck Institute for Biophysical Chemistry
PubChem Primary Bioassay Identifier (AID): AID-1519
Probe Structure & Characteristics
| PubChem CID | CID-310557 |
| Molecular Weight | 273.76 |
| Molecular Formula | C15H12FNO3 |
| XLogP | 3.4 |
| H-Bond Donor | 1 |
| H-Bond Acceptor | 4 |
| Rotatable Bond Count | 4 |
| Exact Mass | 273.08 |
| Topological Polar Surface Area | 55.4 |
| Heavy Atom Count | 20 |

| CID/ML | Target Name | IC50/EC50 (μM) [SID, AID] | Anti-target Name(s) | IC50/EC50 (μM) [SID, AID] | Selectivity | Selectivity Assay(s) Name: IC50/EC50 (nM) [SID, AID]§ |
|---|---|---|---|---|---|---|
| CID-310557/ML084 | COPI | 5 ± 2.5 [SID-11114231, AID-1519] | Cytotoxicity | Inactive @40 μM, [SID-11114231, AID-1561] | >10-fold | Cell-Titer Glo [SID-11114231, AID-1561] |
Recommendations for the scientific use of this probe
The probe Exo-1 is a modulator of protein trafficking. Our work has linked the Golgi Type I coat proteins known as cotamer, COPI to lipid storage. Exo-1 can be used to induce a lipid storage phenotype in cells through modulation of the protein trafficking pathways.
Specific Aim
We have developed a cell-based assay in Drosophila melanogaster embryonic Kc167 and S3 cells capable of analyzing the effects of alterations in lipid metabolism. We have recently used a similar assay successfully in a genome-wide RNAi screen to identify gene functions regulating cellular lipid storage. This assay was adapted to laser-scanning microplate cytometry suitable for a 1,536-well microtiter plate format, which will allow the assay to be screened against the 300K MLSMR collection. The goal is to identify chemical probes affecting lipid storage regulation.
Significance
The chemical probes yielded by this project should be useful tools for providing a better understanding of cellular and organismic lipid storage on a functional and evolutionary level. Furthermore, active substances might result in the identification of lead compounds for the treatment of emerging lipid storage-associated diseases, including atherosclerosis, diabetes or obesity. Also, the present assay will establish a profile of compounds within the MLSMR that modulate this ubiquitous area of biology. The probe described here came from initial screening of known bioactive compound collections. By combining the small molecule screening results with lipid metabolism modulating gene functions identified in the genome-wide RNAi screen, we were able to identify COPI proteins as negative regulators of lipid storage and the probe Exo1 as chemical probe to modulate lipid storage.
Rationale
Storing lipids as a reservoir for energy or the anabolism of elementary metabolites is a common feature of life in organisms from bacteria to humans. The universal cellular lipid storage organelle is the so-called lipid storage droplet (LD). Despite their ubiquitous nature, LDs share a simple, stereotyped structure of a hydrophobic core harboring the storage lipids, which is shielded by a droplet-specific phospholipid monolayer to which proteins are attached. The current model of LD biogenesis involves an incorporation of the lipid core into the membrane leaflets of the endoplasmic reticulum (ER), followed by a subsequent budding-like maturation of a LD, which ultimately pinches off. Once released, LD volume can increase by localized lipogenesis or fusion of existing droplets. Storage lipids are re-mobilized enzymatically by lipase activity. Lipase regulation in the adipocyte is heavily studied and involves multiple components including catecholamine signaling (1), the LD-associated proteins Perilipin and comparative gene identification 58 (CGI-58) (2), and at least two lipases named hormone sensitive lipase (HSL) (3) and adipocyte triglyceride lipase (ATGL) (4–6). LD biogenesis, turnover and mobilization are poorly understood and only few components are known. However, there is an urgent need to learn more about ectopic fat depots as mislocalized storage of lipids, for example in the liver or muscle, is an eminent health problem associated with insulin resistance or the metabolic syndrome (7).
Assay Implementation and Screening
PubChem Bioassay Name
Dmel lipid storage
List of PubChem bioassay identifiers generated for this screening project (AIDs)
Primary Assay Description as defined in PubChem
Overview
The primary goal of this project is to identify chemical probes that either increase or decrease the lipid content of cells. Most cells are capable of storing energy rich lipids (mainly triacylglycerols), which are generated on basis of de novo synthesized fatty acids or non-esterified free fatty acids (NEFA) taken up from the environment. There are only few drugs for treating metabolic diseases and a very limited number of chemical probes to study lipid storage in vitro. We have found that embryonic Drosophila S3 and Kc167 cells are capable of depositing LDs, and we developed an assay to measure lipid storage amounts by fluorescent staining of LDs in S3 cells, using laser-scanning microplate cytometry for detection. Importantly, most results obtained in the Drosophila cell systems could be successfully translated into different mammalian cell culture models, opening up the possibility to test for evolutionary conservation of mechanism.
Protocol
Detection of lipid droplets was performed using Drosophila S3 (obtained from NIDDK) cells, following 400 μM oleic acid feeding in growth media. In the assay 4 μL of S3 cells at 1.25 x 106 cells/mL in growth media was dispensed into LoBase Aurora 1,536-well plates (black walled, clear bottom COP plastic; Aurora) (8) using a bottle-valve solenoid-based dispenser (9) to give 5,000 cells/well. Twenty-three nL of compound solution was transferred to the assay plates using a Kalypsys pin tool (San Diego, CA) equipped with a 1,536-pin array containing 10 nL slotted pins (FP1S10, 0.457 mm diameter, 50.8 mm long; V&P Scientific5). Next, 1 μl of oleic acid (400 μM) was added and the plates were lidded with stainless steel rubber gasket-lined lids containing pin-holes for gas exchange and incubated for 18–24 hr at 24 °C, 95% humidity. Detection was performed by the addition of the lipid-droplet specific dye BODIPY 493/503 (Molecular Probes), and CellTracker™ Red CMTPX (Invitrogen) was used to enumerate cell number. Lipid droplet accumulation was measured using the Acumen Explorer (TTP LapTech) (10) using a 488 nm laser. The total intensity in channel 1 (500–530 nm) was used to measure lipid droplet accumulation with cell objects defined using channel 3 (575–640 nm) and a 5 μm width and 100 μm depth filters. The ratio of the total intensity in PMT channel 1 over total intensity channel 3 was also calculated.
Summary of the Primary Screen
Assay principle and protocol

Figure 1
Top, distribution of the fluorescent signal in PMT 1 (lipid specific channel) without (left) and with (right) oleic acid (OA) feeding. The populations are defined using width and depth filters using the fluorescent signal from PMT 3 (cell- specific channel) to define cell-objects. Cell objects with low PMT 1 fluorescence are defined in the absence of OA feeding (“unfed population”, orange data), and a shift to brighter fluorescence is observed following OA feeding that is used to define lipid-containing cells (“fed population”, green data at top right). Unclassified objects not used in the calculation are shown as red. Middle pictures show false color (same color code as in histograms) plate well images from the Acumen for unfed (left) and fed (right) cells. The bottom two panels represent images from the InCell1000 using the same cell stains. Cells are treated with 400 μM OA in the presence (top) or absence (bottom) of the inhibitor Triacsin C. In the fused image, the InCell software swaps the colors so that the red color represent the lipid stain and the green represent the cell stain.
The optimized 1536-well protocol is given in Table 1.
Table 1
Final 1536-well assay protocol.
Following adaptation of the lipid droplet assay to 1536-well plates, we used the assay to screen several libraries. The libraries were screened at between seven and fifteen concentrations using quantitative HTS (qHTS) (11). A total of 98 1536-well plates were screened (Figure 2). This included 27 DMSO blanks, a 15 point titration of the LOPAC library (Sigma), a 13 point titration of the Prestwick library, seven point titrations of two combinatorial libraries targeted to GPCRs and kinases (PTL1 and PTL2), seven point titrations of two libraries from Spectrum/Micorsource and a fifteen point titration of the Tocris library. Overall the assay showed excellent performance with a Z′ = 0.77 ± 0.23, S/B = 25 ± 68, CV = 3 ± 3%. Three plates failed QC (arrows and low Z-factor plates in Figure 2c,d) due to a clogged tip.
Identification of Exo1
Following the qHTS, the CRC data was subjected to a classification scheme to rank the quality of the CRCs as described by Inglese and co-workers (11) (see scheme 1). Briefly, CRCs are placed into four classes. Class 1 contains complete CRCs showing both upper and lower asymptotes and r2 values > 0.9. Class 2 contains incomplete CRCs lacking the lower asymptote and shows r2 values greater than 0.9. Class 3 curves are of the lowest confidence because they are defined by a single concentration point where the minimal acceptable activity is set at 3 SD of the mean activity calculated from the lowest tested concentration. Finally, class 4 contains compounds that do not show any CRCs and are therefore classified as inactive.
qHTS summary of assay results
Compounds showing decreasing lipid storage were generally associated with cytotoxicity, except for the positive control Triacsin C as identified in the red cell stain channel. However, we found many compounds that induced lipid overstorage (Figure 3). In the Tocris library, we noted a small molecule Arf1 modulator (Exo1) (12) that increased lipid droplet accumulation (EC50 = 5 μM) in S3 cells (Figure 4). This compound is known to inhibit the vesicle-mediated Coat Protein complex I (COPI) transport complex. Further, we have identified key Drosophila candidate genes for lipid droplet regulation by RNA interference (RNAi) screening that included the COPI transport complex, which was found to be required for limiting lipid storage.
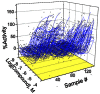
Figure 3
Example CRCs (fed objects) for lipids storage activators from the qHTS of S3 cells.
Probe Characterization
We found that interference with COPI function by RNAi in Drosophila Kc167 cells, as well as in mouse 3T3-L1 or AML12 cells, results in increased lipid storage (see Figure 4 and Beller et al., (13)). In order to characterize the COPI knockdown effects on lipid storage in greater detail, and to confirm the independently identified effect of the Exo1 compound, we utilized different secondary assays. In addition to Exo1, we used BFA, another compound that has been implicated in the modulation of COPI-mediated trafficking in mammalian cells (14). In order to support our finding of evolutionary conservation of COPI effects on lipid storage, we also utilized Exo1 and BFA in the mammalian cell system. Both compounds reduced NEFA release to the same extent as the siRNAs targeting COPI subunit encoding mRNAs (Figure 5 and see Ref [13]). Mimicking genetic epistasis experiments by combining RNAi and compound treatment, we furthermore obtained results supporting the hypothesis that COPI-functions in the same pathway as ATGL and might be involved in its activation (see Figure 5 and Ref[13]), opening up new possibilities to study this currently heavily studied question. Positive regulation of lipolysis by the COPI retrograde vesicle trafficking pathway was the most striking and unexpected result of both the RNAi and small molecule compound screen. We propose that COPI is likely to function directly at the lipid droplet surface and rather than indirectly through the Golgi. See Beller et al. (13) for a full description of these findings.
Mode of action for Exo1
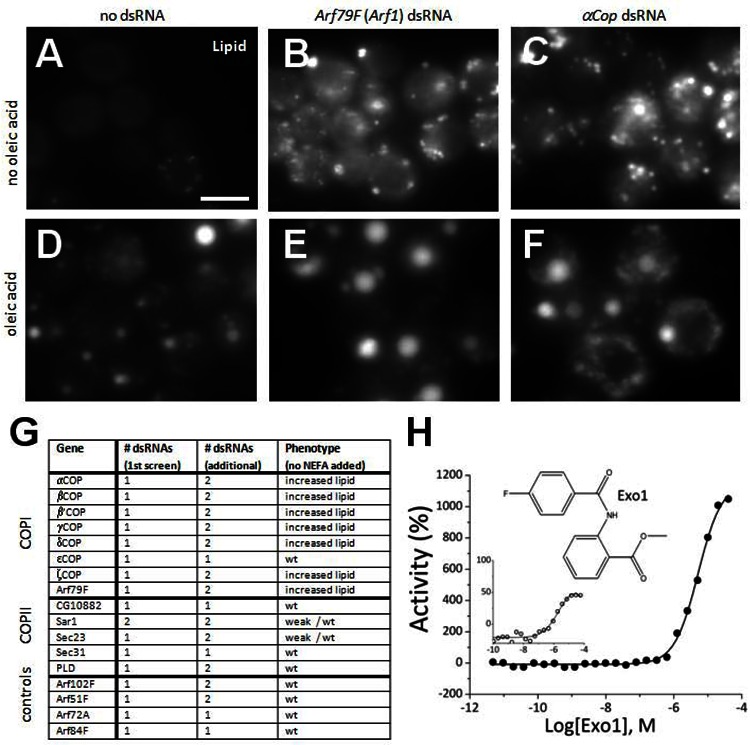
Figure 4The COPI-Mediated Retrograde Trafficking Pathway is a negative regulator of lipid storage
(A–F) Drosophila cells with or without oleic acid stained with BODIPY493/503 to detect lipid. Control cells not treated with dsRNA (A and D) or cells incubated with dsRNAs targeting Arf79F (an Arf1 homolog) (B and E) or alphaCop (C and F) are shown. (G) All COPI, and several COPII members as well as additional Arfs, were retested using independent dsRNAs and gave similar results. Results and number of dsRNAs present in the primary screen (including oleic acid) and retests are given. (H) Dose response of Drosophila S3 cells to Exo1 (structure inset) showing the %-activity derived either from the lipid specific signal (filled circles) or the lipid/cell ratio (open circles). Percent activity refers hereby to the changes of lipid storage relative to Triacsin C treatment, which decreases lipid storage by blocking TG synthesis. Increased activity indicates increased lipid storage, which increased with concentration. Scale bar in (A) represents 10. Adopted from Beller et al. 2008. (13)

Figure 5NEFA Incorporation and NEFA Release Measured in AML12 Cells after compound treatment
Relative activity [(experimental/ALLStars negative control (control) and DMSO) Radiolabel assays for NEFA release (nM) relative to total protein concentration in cells treated with siRNAs targeting the indicated transcripts in the presence of DMSO only (open bar), BFA (5 μM) in DMSO (light-grey bar), or Exo1 (5 μM) in DMSO (dark-grey bar). Significance at p<0.01, unpaired t-test, is shown (indicated by an asterisk [*]). Standard error is indicated by the bars.
Synthesis of Exo1 analogs
General procedure for the synthesis of Exo-1 (CID-310557) and its analogues
To a solution of the appropriate aryl acid (220 mg, 1.45 mmol) in DCM (20 ml) was added SOCl2 (2.12 ml, 29.0 mmol) and several drops of DMF, and the mixture was stirred at room temperature for 0.5 h. The solvent and excess SOCl2 were removed under reduced pressure and the resulting film was reconstituted in DCM (10 ml) followed by the addition of Et3N (2.42 ml, 1.74 mmol, 1.2 eq.) and the appropriate analine (253 mg, 1.60 mmol, 1.1 eq). The resulting mixture was stirred for 3 h and water (10 ml) was added, and the organic layer was separated. The aqueous layer was further extracted with DCM (20 ml). The combined organic layers were washed with brine and dried over Na2SO4. After the removal of solvent, the residue was purified by column chromatography (EtOAc/DCM= 1/40) to afford desired product (297 mg, 75%).

1H NMR (400 MHz, CDCl3) δ 12.03 (brs, 1H), 8.92 (dd, JHH= 8.6, 1.0 Hz, 1H), 8.04–8.15 (m, 3H), 7.61 (td, JHH= 8.0, 1.6 Hz, 1H), 7.20–7.43 (m, 3H), 3.97 (s, 3H); (TOFMS) m/z 274.0878 (M+H+) (calculated for C15H13FNO3+) 274.0879.

1H NMR (400 MHz, CDCl3) δ 8.71 (brs, 1H), 8.60 (dd, JHH= 8.0, 1.6 Hz, 1H), 8.42 (d, JHH= 0.4 Hz, 1H), 7.96–8.02 (m, 3H), 7.87–7.92 (m, 1H), 7.55–7.62 (m, 2H), 7.03–7.14 (m, 2H), 6.95 (dd, JHH= 8.0 Hz, 1.6 Hz, 1H), 3.95 (s, 3H); (TOFMS) m/z (M+H+) 278.1196 (calculated for C18H16NO2+) 278.1181

1H NMR (400 MHz, CDCl3) δ 12.21 (brs, 1H), 8.89 (d, JHH= 8.2 Hz, 2H), 8.15 (d, JHH= 8.4 Hz, 1H), 8.11 (dd, JHH= 8.0, 1.6 Hz, 1H), 7.82 (d, JHH= 8.2 Hz, 2H), 7.63 (td, JHH= 7.8, 1.6 Hz, 1H), 7.17 (m, 1H), 3.98 (s, 3H); (TOFMS) m/z 281.0925 (M+H+) (calculated for C16H13N2O3+) 281.0926.

1H NMR (400 MHz, CDCl3) δ 11.82 (d, JHF= 7.2 Hz, 1H), 8.92 (dd, JHH= 8.4, 0.8 Hz, 1H), 8.04–8.15 (m, 2H), 7.60 (td, JHH= 7.0, 1.6 Hz, 1H), 7.14 (td, JHH= 7.6, 0.8 Hz, 1H), 6.90–7.06 (m, 2H), 3.94 (s, 3H); (TOFMS) m/z 292.0781 (M+H+) (calculated for C15H12F2NO3+) 292.0785.
SAR of Exo1 analogs

NCGC-000168461 shows the weakest activity in agreement with the SAR derived from characterization of Exo1 analogs in mammalian cells by Feng et al. (12)
This compound has been provided to the MLSMR: MLS-000722799
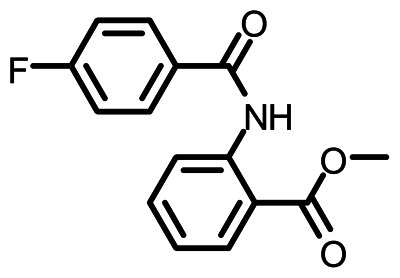
Canonical SMILES: COC(=O)C1=CC=CC=C1NC(=O)C2=CC=C(C=C2)F
InChI: InChI=1/C15H12FNO3/c1-20-15(19)12-4-2-3-5-13(12)17-14(18)10-6-8-11(16)9-7-10/h2-9H,1H3,(H,17,18)/f/h17H
Description of secondary assays used in probe characterization
Lipolysis and lipogenesis measurements in AML12 cells
Measurements of NEFA released from lipid droplets or incorporated into the TG fraction were performed as previously described (13). Briefly, AML12 cells treated with or without specific siRNAs (10 nM) for 4 d were incubated overnight with growth medium, supplemented with 400 μM oleic acid complexed to 0.4% bovine serum albumin, to promote triacylglycerol deposition and [3H] oleic acid, at 1 x 106 dpm/well, was included as a tracer. In lipolysis experiments, re-esterification of fatty acids in AML12 cells was prevented by including 10 μM Triacsin C (Biomol), an inhibitor of acyl coenzyme A synthetase, in the medium. Quadruplicate wells were tested for each condition. Lipolysis was determined by measuring radioactivity released into the media in 1 h. For the lipid extraction and thin layer chromatography, the cell monolayer was washed with ice-cold PBS and scraped into 1 ml of PBS. Lipids were extracted by the Bligh-Dyer method [84], and 10% of the total lipid was analyzed by thin layer chromatography. AML12 cells treated with or without specific siRNAs were additionally incubated with either vehicle (DMSO), 5 μM of Exo1 (12.5 mg/ml DMSO), or BFA (10 mg/ml DMSO) during the time of radioactivity release into the media (2 h). NEFA incorporation into the TG fraction and NEFA release are calculated as nanomoles/milligram protein. Protein measurements were performed using a commercial BCA assay kit (Pierce Biotechnology) according to the manufacturer’s instructions. Statistical significance was tested by impaired Student t test (GraphPad software).
Probe
a. Chemical name
Methyl 2-(4-fluorobenzamido)benzoate (ML084)
b. Probe chemical structure
c. Structural Verification Information of probe SID (11114231)
1H NMR (400 MHz, CDCl3) δ 12.03 (brs, 1H), 8.92 (dd, JHH= 8.6, 1.0 Hz, 1H), 8.04–8.15 (m, 3H), 7.61 (td, JHH= 8.0, 1.6 Hz, 1H), 7.20–7.43 (m, 3H), 3.97 (s, 3H); (TOFMS) m/z 274.0878 (M+H+) (calculated for C15H13FNO3+) 274.0879.
d. PubChem CID (corresponding to the SID)
310557
e. Availability from a vendor
Exo1 is sold by Tocris Biosciences (Tocris-1850).
f. Mode of action for biological activity of probe
Increases lipid droplet storage by inhibition of COPI function.
g. Detailed synthetic pathway for making probe
See page 11
h. Summary of probe properties
Exo1 is not fluorescent using 488 nm excitation. Possess adequate solubility for in vitro use. Probe is not toxic to cells at 60 µM testing concentration (24 hr incubation). Probe properties such as solubility are available at Tocris Biosciences.
Compound preparation
Compound is prepared in DMSO at 10 mM stock concentration. Assays described above have 0.6% DMSO final concentration in buffer.
Bibliography
- 1.
- Langin D. Adipose tissue lipolysis as a metabolic pathway to define pharmacological strategies against obesity and the metabolic syndrome. Pharmacol Res. 2006;53:482–91. [PubMed: 16644234]
- 2.
- Subramanian V, Rothenberg A, Gomez C, Cohen AW, Garcia A, Bhattacharyya S, Shapiro L, Dolios G, Wang R, Lisanti MP, Brasaemle DL. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J Biol Chem. 2004;279:42062–71. [PubMed: 15292255]
- 3.
- Vaughan M, Berger JE, Steinberg D. Hormone-Sensitive Lipase and Monoglyceride Lipase Activities in Adipose Tissue. J Biol Chem. 1964;239:401–9. [PubMed: 14169138]
- 4.
- Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, Jackle H, Kuhnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–30. [PubMed: 16054079]
- 5.
- Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, Heldmaier G, Maier R, Theussl C, Eder S, Kratky D, Wagner EF, Klingenspor M, Hoefler G, Zechner R. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–7. [PubMed: 16675698]
- 6.
- Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, Zechner R. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–6. [PubMed: 15550674]
- 7.
- Frayn KN, Arner P, Yki-Jarvinen H. Fatty acid metabolism in adipose tissue, muscle and liver in health and disease. Essays Biochem. 2006;42:89–103. [PubMed: 17144882]
- 8.
- Niles WD, Coassin PJ. Cyclic olefin polymers: innovative materials for high-density multiwell plates. Assay Drug Dev Technol. 2008;6:577–90. [PubMed: 18537466]
- 9.
- Niles WD, Coassin PJ. Piezo- and Solenoid Valve-Based Liquid Dispensing for Miniaturized Assays. Assay Drug. Devel. Technol. 2005;3:189–202. [PubMed: 15871693]
- 10.
- Bowen WP, Wylie PG. Application of Laser-Scanning Fluorescence Microplate Cytometry in High Content Screening. Assay Drug Dev Technol. 2006;4:209–221. [PubMed: 16712425]
- 11.
- Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103:11473–8. [PMC free article: PMC1518803] [PubMed: 16864780]
- 12.
- Feng Y, Yu S, Lasell TK, Jadhav AP, Macia E, Chardin P, Melancon P, Roth M, Mitchison T, Kirchhausen T. Exo1: a new chemical inhibitor of the exocytic pathway. Proc Natl Acad Sci U S A. 2003;100:6469–74. [PMC free article: PMC164470] [PubMed: 12738886]
- 13.
- Beller M, Sztalryd C, Southall N, Bell M, Jackle H, Auld DS, Oliver B. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 2008;6:2530–2549. [PMC free article: PMC2586367] [PubMed: 19067489]
- 14.
- Lippincott-Schwartz J, Yuan L, Tipper C, Amherdt M, Orci L, Klausner RD. Brefeldin A’s effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell. 1991;67:601–16. [PubMed: 1682055]
- PMCPubMed Central citations
- PubChem SubstanceRelated PubChem Substances
- PubMedLinks to PubMed
- Review Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.[Brain Neurotrauma: Molecular, ...]Review Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.Wolahan SM, Hirt D, Glenn TC. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. 2015
- Review Modulators of Lipid Storage.[Probe Reports from the NIH Mol...]Review Modulators of Lipid Storage.Boxer MB, Shen M, Zhang Y, Liu L, Auld DS, Beller M. Probe Reports from the NIH Molecular Libraries Program. 2010
- Genome-wide screens for gene products regulating lipid droplet dynamics.[Methods Cell Biol. 2012]Genome-wide screens for gene products regulating lipid droplet dynamics.Fei W, Yang H. Methods Cell Biol. 2012; 108:303-16.
- Phenotypic screening with oleaginous microalgae reveals modulators of lipid productivity.[ACS Chem Biol. 2013]Phenotypic screening with oleaginous microalgae reveals modulators of lipid productivity.Franz AK, Danielewicz MA, Wong DM, Anderson LA, Boothe JR. ACS Chem Biol. 2013 May 17; 8(5):1053-62. Epub 2013 Mar 22.
- Review A novel strategy to identify drugs that interfere with endosomal lipids.[Chimia (Aarau). 2011]Review A novel strategy to identify drugs that interfere with endosomal lipids.Moreau D, Scott C, Gruenberg J. Chimia (Aarau). 2011; 65(11):846-8.
- Identification of lipid storage modulators - Probe Reports from the NIH Molecula...Identification of lipid storage modulators - Probe Reports from the NIH Molecular Libraries Program
Your browsing activity is empty.
Activity recording is turned off.
See more...


