NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
Placental alkaline phosphatase (PLAP) is highly expressed in primate placental tissue. Its biological function and relevance are still unknown, but PLAP-like enzymes are detected in serum of patients with primary testicular tumors, in particular seminoma and other cancers. Consequently, the identification of PLAP-specific inhibitors with selectivity over tissue non-specific alkaline phosphatase (TNAP) and intestinal alkaline phosphatase (IAP) may provide the necessary tools to characterize its biological role. Currently, inhibitors of PLAP lack either potency or selectivity. The small molecule probe ML085 (CID-25067483) is a biochemical inhibitor of PLAP, and will be useful to elucidate the key biological functions and natural substrates of human PLAP.
Assigned Assay Grant #: 1 R03 MH077602-01
Screening Center Name & PI: Conrad Prebys Center for Chemical Genomics (formerly Burnham Center for Chemical Genomics) & Dr. John C. Reed
Chemistry Center Name & PI: Conrad Prebys Center for Chemical Genomics (formerly Burnham Center for Chemical Genomics) & Dr. John C. Reed
Assay Submitter & Institution: Dr José Luis Millán & Sanford-Burnham Medical Research Institute (formerly Burnham Institute for Medical Research)
PubChem Summary Bioassay Identifier (AID): AID-1577
Probe Structure & Characteristics

| CID/ML | Target Name | IC50/EC50 (nM) [SID, AID] | Anti-target Name(s) | IC50/EC50 (μM) [SID, AID] | Selectivity | Secondary Assay(s) Name: IC50/EC50 (nM) [SID, AID] |
|---|---|---|---|---|---|---|
| CID-665093 ML085 | PLAP | 2,600 nM IC50 SID-56373725 AID-1512 | TNAP | >100 µM IC50 SID-56373725 AID-518 | > 42 | |
| IAP | >6.25 µM IC50 SID-56373725 AID-1017 | >2.4 |
Recommendations for the scientific use of this probe
Placental alkaline phosphatase (PLAP) is highly expressed in primate placental tissue. Its biological function is still unknown (1,2). PLAP-like enzymes could be detected in serum of patients with primary testicular tumors, in particular seminoma (3) and other cancers (4). This PLAP-specific inhibitors with selectivity over tissue non-specific alkaline phosphatase (TNAP) and intestinal alkaline phosphatase (IAP) can be used as a tools to characterize the biological role of PLAP. The small molecule probe CID-665093 will be useful to elucidate the key biological functions and natural substrates of human placental alkaline phosphatase (PLAP)
1. Scientific Rationale for Project
Alkaline phosphatases (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified: three isozymes are tissue-specific and the fourth one is tissue-nonspecific. Placental alkaline phosphatase (PLAP) is highly expressed in primate placental tissue. Its biological function is still unknown. However, the identification of PLAP-specific inhibitors with selectivity over tissue non-specific alkaline phosphatase (TNAP) and intestinal alkaline phosphatase (IAP) will provide the necessary tools to characterize its biological role
2. Project Description
a. The original goal for probe characteristics
This MLSCN carry-forward project was an early Cycle 2 assay proposal and a formal CPDP was not filed. It was derived on work with tissue non-specific alkaline phoshatase inhibitor and later activator work, that suggested that PLAP specific inhibitors would also useful tools for this class of isozyme with yet to be elucidated biological function.
b. Assay implementation and screening
i. PubChem Bioassay Name(s), AID(s), Assay-Type (Primary, DR, Counterscreen, Secondary)
| PubChem BioAssay Name | AIDs | Probe Type | Assay Type | Assay Format | Assay Detection & well format |
|---|---|---|---|---|---|
| Luminescent assay for HTS discovery of chemical inhibitors of placental alkaline phosphatase | 690 | Inhibitor | Primary | Biochemical | Luminescence (1536) |
| Luminescent assay for HTS discovery of chemical inhibitors of placental alkaline phosphatase confirmation | 1512 | Inhibitor | Confirmatory | Biochemical | Luminescence (384) |
| TNAP luminescent HTS assay [Confirmatory] | 518 | Inhibitor | Secondary Assay for specificity | biochemical | Luminescence (384) |
| Luminescent assay for identification of inhibitors of human intestinal alkaline phosphatase [Confirmatory] | 1017 | Inhibitor | Counterscreen Assay for specificity | biochemical | Luminescence (1536) |
| Luminescent assay for identification of inhibitors of bovine intestinal alkaline phosphatase [Primary Screening] | 1019 | Inhibitor | Counterscreen Assay for specificity | biochemical | Luminescence (1536) |
Primary assay details are described below.
PLAP screening was developed and performed at the Burnham Center for Chemical Genomics (BCCG) within the Molecular Library Screening Center Network (MLSCN) as a selectivity screen for tissue nonspecific alkaline phosphatase (TNAP, AID-518). XO1 MH077602-01, Pharmacological inhibitors of tissue-nonspecific alkaline phosphatase (TNAP), Assay Provider Dr. José Luis Millán, Burnham Institute for Medical Research, La Jolla, CA.
Protocol
PLAP assay materials
- PLAP protein was provided by Dr. José Luis Millán (Burnham Institute for Medical Research, San Diego, CA). The CDP-star was obtained from New England Biolabs.
- Assay Buffer: 250 mM DEA, pH 9.8, 2.5 mM MgCl2, and 0.05 mM ZnCl2.
- PLAP working solution contained a 1/6400 dilution in assay buffer. The solution was prepared fresh prior to use.
- CDP-star working solution contained 212.5 µM CDP-star in MQ water.
- TCEP working solution −5 mM in 10% DMSO.
PLAP HTS protocol
- 4 uL of 100 µM compounds in 10% DMSO were dispensed in columns 3–24 of Greiner 384-well white small volume plates (784075).
- Using a Thermo wellmate dispenser 4 uL of the following solutions were added:
- TCEP working solution - column 1 (positive control).
- 10% DMSO - column 2 (negative control).
- 8 uL of PLAP working solution was added to the whole plate using a WellMate bulk dispenser (Matrix).
- 8 uL of CDP-star working solution was added to the whole plate using WellMate bulk reagent dispenser (Matrix).
- Final concentrations of the components in the assay were as follows:
- 100 mM DEA, pH 9.8, 1.0 mM MgCl2, 0.02 mM ZnCl2 (columns 1–24)
- 1/16000 dilution PLAP (columns 1–24)
- 85 µM CDP-star (columns 1–24)
- 1 mM TCEP (columns 1)
- 2 % DMSO (columns 1–24)
- 20 µM compounds (columns 3–24)
- Plates were incubated for 30 minutes at room temperature.
- Luminescence was measured on an Envision plate reader (Perkin Elmer).
- Data analysis was performed using CBIS software (ChemInnovations, Inc).
PLAP dose-response confirmation screening protocol
- Dose-response curves contained 10 concentrations of compounds obtained using 2-fold serial dilution. Compounds were serially diluted in 100% DMSO, and then diluted with water to 10% final DMSO concentration. 4 uL compounds in 10% DMSO were transferred into columns 3–22 of Greiner 384-well white small-volume plates (784075). Columns 1–2 and 23–24 contained 4 uL of TCEP working solution and 10% DMSO, respectively.
- 8 uL of PLAP working solution was added to the whole plate using a WellMate bulk reagent dispenser (Matrix).
- 8 uL of CDP-star working solution was added to the whole plate using a WellMate reagent bulk dispenser (Matrix).
- Plates were incubated for 30 mins at room temperature.
- Luminescence was measured on an Envision plate reader (Perkin Elmer).
- Data analysis was performed using CBIS software (ChemInnovations, Inc) using a sigmoidal dose-response equation through non-linear regression
ii. Assay Rationale & Description
For the assay rationale, description and reagents protocols see section 2a above.
For this screen 95857 compounds were tested. The average Z′ for the assay was 0.69, the average signal to background was 38.6, the average signal to noise was 122.7 and the average signal window was 7.8. Initially 192 compounds were identified as primary positives with >= 50% inhibition of activity in the assay. After performing dose-response experiments with liquid DMSO stocks of these compounds, 82 of the hits generated a dose-response curve.
iii. Summary of Results
Primary hits from the PLAP HTS assay were confirmed and tested in parallel against TNAP for selectivity. A few selective hits were identified. Those hits were purchased along with 56 analogues (Table 1). There are many commercial analogues available and so Analogue-By-Catalogue (ABC) was performed in an iterative fashion. From the purchased compounds we were able to conclude that the dihydroxyl groups of the catechol are essential for PLAP activity. The SAR was completed and a probe molecule CID-665093 (MLS-0014097) was identified (see Table 1 below).
Table 1
SAR results generated using analogue-by-catalogue (ABC).
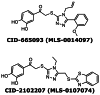
Medicinal chemistry focused on the two final probe candidates, CID-2102207 (MLS-0107074) and CID-665093 (MLS-0014097) (Figure 1) that were identified as low-micromolar inhibitors of PLAP through primary screening.
Re-synthesis of these targets was undertaken and these compounds were subjected to the BCCG target independent ADME assay panel (see also Table 1 below)
c. Probe Optimization
Description of SAR & chemistry strategy (including structure and data) that led to the probe
SAR was developed primarily by the ABC approach for this MLSCN project due to the availability of numerous commercial analogues. Analysis of the SAR identified some key structural features, including: (1) Reposition or derivatization of the hydroxyl groups leads to inactive compounds (SID-56373705, SID-56373700 and SID-56373696). (2) Selectivity for PLAP apparently resides in the N-substituted triazole group (Figure 1). Unsubstituted triazoles also potently inhibit PLAP but do not display selectivity vs. TNAP and IAP. Though 58 analogues were purchased to explore R1 and R2 (see Figure 2, and attached triazole list at end), no significant improvement in either selectivity or potency was achieved. For example, analogue SID-57287606 is in the submicromolar range for PLAP, but it is not as selective as the proposed probe molecule. The objective of chemistry was to develop a tractable route for the synthesis of the probe candidates, prepare authentic samples, confirm structure and purity, and provide enough material for submission to MLSMR.
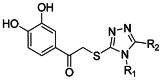
Figure 2
Probe analogs R1 & R2 variations.
3. Probe
a. Chemical name
1-(3,4-dihydroxyphenyl)-2-[[5-(2-methoxyphenyl)-4-prop-2-enyl-1,2,4-triazol-3-yl]sulfanyl]ethanone [ML085]
b. Probe chemical structure
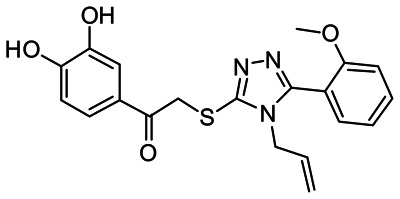
Figure 3Probe compound structure
c. Structural Verification Information of probe SID
Spectral data supporting proposed structure: 1H NMR (400 MHz, Acetone-D6) δ 7.54 (ddd, J = 8.4, 7.5, 1.8, 1H), 7.46 – 7.33 (m, 3H), 7.17 (d, J = 8.1, 1H), 7.07 (dt, J = 7.5, 0.9, 1H), 6.72 (d, J = 8.2, 1H), 5.89 – 5.60 (m, 1H), 5.10 (dd, J = 10.4, 1.1, 1H), 4.91 (dd, J = 17.2, 1.1, 1H), 4.78 (s, 2H), 4.49 (dd, J = 3.7, 1.6, 2H), 3.83 (s, 3H). 13C NMR (101 MHz, Acetone-D6) δ 191.32, 159.49, 158.26, 154.41, 151.21, 148.89, 133.16, 132.97, 132.87, 124.45, 123.57, 121.53, 117.97, 117.73, 115.86, 113.80, 112.18, 55.92, 47.44, 42.01. MS (ESI) calculated C20H19N3O4S m/z = 397.11, found m/z = 398.01 [M+H].
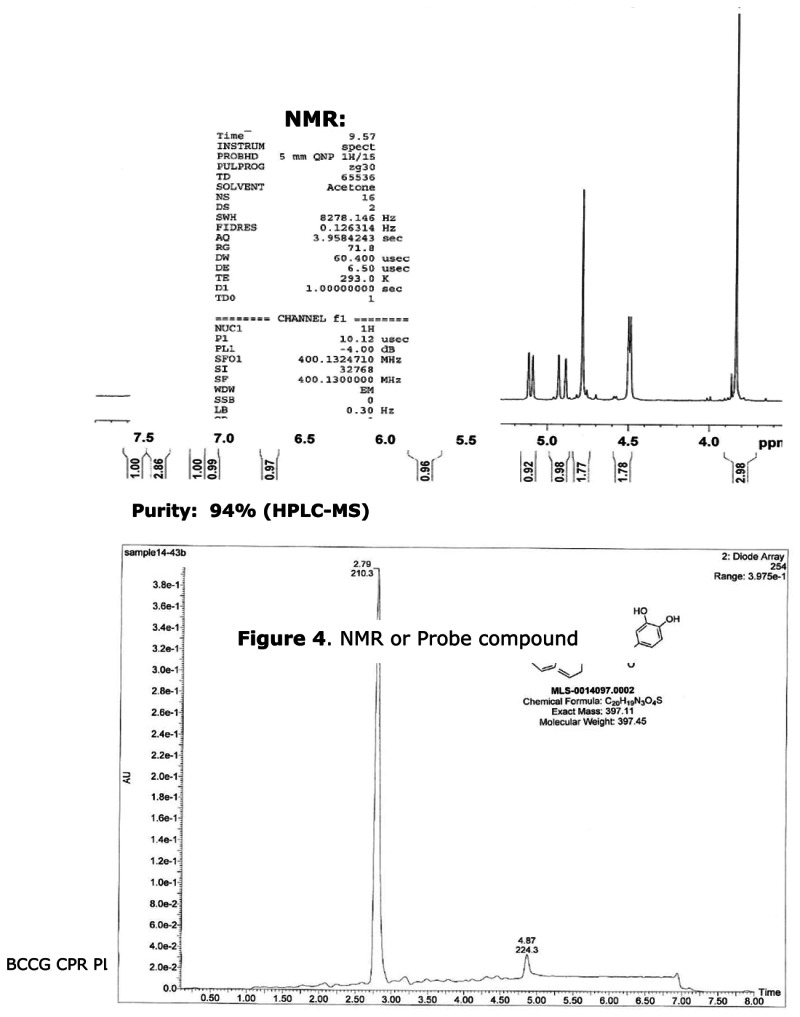
Figure 4NMR or Probe compound
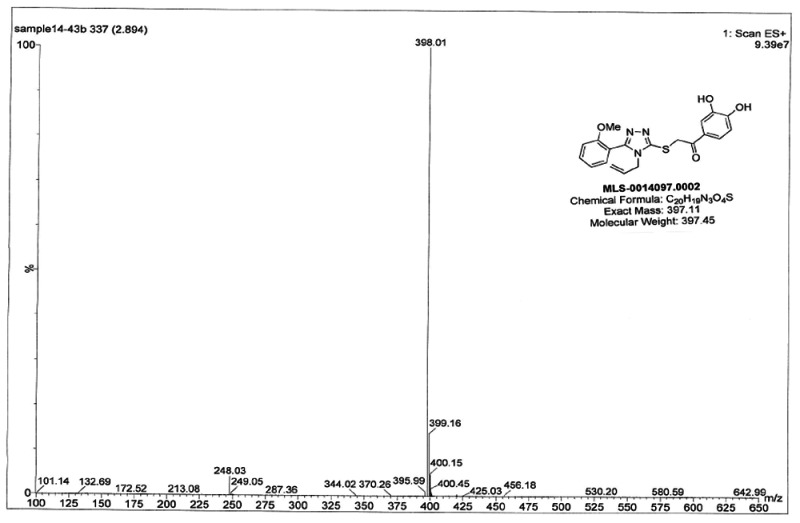
Figure 5LC-MS traces for probe purity
d. PubChem CID (corresponding to the SID)
CID-665093
e. Availability from a vendor
The probe is commercially available from InterBioScreen (ID # STOCK4S-90308).
f. MLS# that verifies the submission of probe molecule and five related samples that were submitted to the SMR collection
Table 2Submission information on Probe and analogs
| Probe/Analog | MLS-# (BCCG#) | CID | SID | Source (vendor or BCCG syn) | Amt (mg) | Date ordered/submitted |
|---|---|---|---|---|---|---|
| Probe | 0014097 | 665093 | 56373725 | InterBioScreen | 20 | 4/15/09 |
| Analog 1 | 0390843 | 1409759 | 57287589 | InterBioScreen | 20 | 4/15/09 |
| Analog 2 | 0390850 | 1400698 | 57287596 | InterBioScreen | 20 | 4/15/09 |
| Analog 3 | 0390851 | 1400246 | 57287597 | InterBioScreen | 20 | 4/15/09 |
| Analog 4 | 0390857 | 1398345 | 57287604 | InterBioScreen | 20 | 4/15/09 |
| Analog 5 | 0390858 | 1394536 | 57287605 | InterBioScreen | 20 | 4/15/09 |
g. Mode of action for biological activity of probe
The probe is a biochemical inhibitor of PLAP. The mode of action for the biological activity of this probe has not yet been elucidated.
h. Detailed synthetic pathway for making probe
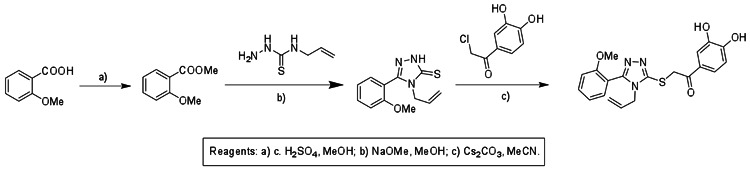
Solid 2-methoxybenzoic acid (5.0g, 32.9mmol) was transferred to a 250ml round-bottom flask and dissolved in 50 mL dry MeOH. 8 drops of c.H2SO4 were added to the reaction mixture. The reaction mixture was heated at reflux for 12h. After 12 h the reaction mixture was cooled to room temperature and the solvent was removed under reduced pressure. The crude reaction mixture was dissolved in 50 mL diethylether and washed with saturated NaHCO3 (2 × 50mL) and brine solution (2 × 50mL). The organic layer was collected and the solvent was removed under reduced pressure to yield the methyl ester as a white solid (5.4 g, 99%). 1H NMR (400 MHz, CDCl3) δ 7.82 (dd, J = 7.9, 1.8, 1H), 7.49 (ddd, J = 8.5, 7.4, 1.8, 1H), 7.00 (dtd, J = 4.7, 3.4, 1.0, 2H), 3.93 (s, 3H), 3.91 (s, 3H).
Methyl anisate (50.0mg, 0.3 mmol) was transferred to a glass vial and dissolved in 5 mL dry MeOH. 4-Allyl thio-semicarbazide (59.2 mg, 0.45 mmol) was added to the reaction mixture. This was followed by addition of 0.2 mL of 25%(wt) NaOMe solution. The resultant reaction mixture was heated overnight at 85oC in a sealed vial. After the reaction was complete, the solvents were evaporated under reduced pressure. The reaction mixture was cooled to 0oC and 20 mL deionized water was added to it. The pH of the reaction mixture was lowered to 5 using 10% AcOH (dropwise addition). The resultant white precipitate (4-allyl-3-(2-methoxyphenyl)-1H-1,2,4-triazole-5(4H)-thione) was filtered, dried and weighed (55mg, 74.3%). 1H NMR (400 MHz, CDCl3) δ 7.65 – 7.47 (m, 1H), 7.35 (dd, J = 7.5, 1.6, 1H), 7.08 (ddd, J = 19.5, 13.1, 4.6, 2H), 5.75 (ddd, J = 16.0, 11.4, 5.7, 1H), 5.10 (d, J = 10.3, 1H), 4.91 (d, J = 17.1, 1H), 4.60 (d, J = 5.8, 2H), 3.86 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 167.79, 157.60, 150.56, 133.03, 131.79, 130.63, 120.98, 118.47, 114.67, 111.15, 55.58, 46.99.
Solid 4-allyl-3-(2-methoxyphenyl)-1H-1,2,4-triazole-5(4H)-thione (25.0 mg, 0.1mmol) was dissolved in 5 mL dry acetonitrile. 2-Chloro-3′,4′-dihydroxyacetophenone (18.8mg, 0.1mmol) was added to the reaction mixture followed by addition of Cs2CO3( 33mg, 0.1mmol). the resultant reaction mixture was stirred at room temperature for 8 hrs. The solid was filtered from the reaction mixture and the filtrate was concentrated to yield the final product 2-(4-allyl-5-(2-methoxyphenyl)-4H-1,2,4-triazol-3-ylthio)-1-(3,4-dihydroxyphenyl)ethanone (25mg, 62.5%). The product was further purified by crystallization using acetone. 1H NMR (400 MHz, Acetone-D6) δ 7.54 (ddd, J = 8.4, 7.5, 1.8, 1H), 7.46 – 7.33 (m, 3H), 7.17 (d, J = 8.1, 1H), 7.07 (dt, J = 7.5, 0.9, 1H), 6.72 (d, J = 8.2, 1H), 5.89 – 5.60 (m, 1H), 5.10 (dd, J = 10.4, 1.1, 1H), 4.91 (dd, J = 17.2, 1.1, 1H), 4.78 (s, 2H), 4.49 (dd, J = 3.7, 1.6, 2H), 3.83 (s, 3H). 13C NMR (101 MHz, Acetone-D6) δ 191.32, 159.49, 158.26, 154.41, 151.21, 148.89, 133.16, 132.97, 132.87, 124.45, 123.57, 121.53, 117.97, 117.73, 115.86, 113.80, 112.18, 55.92, 47.44, 42.01. MS (ESI) calculated C20H19N3O4S m/z = 397.11, found m/z = 398.01 [M+H].
i. Summary of probe properties (solubility, absorbance/fluorescence, reactivity, toxicity, etc.)
Our internal counter screen efforts has showed CID-665093 to be inactive in two assays TNAP inhibition (AID-518) and G6DPH (AID-1020), and 10-fold less active vs. IAP (AID-1017), a very close family member of PLAP.
In Vitro Pharmacology
Compounds CID-665093 (MLS-0014097) and CID-2102207 (MLS-0107074) were subjected to a battery of in vitro pharmacology assays to assess aqueous solubility, cellular permeability, plasma protein binding (PPB), plasma stability, and microsome stability. The results of these assays are presented in Table 2.
Table 3Pharmacologic Properties of MLS-01014097, MLS-0107074
| Cpd-ID | Aqueous Solubility (μg/mL) pH 5.0/6.2/7.4 (AID-1615) | Permeability (×10−6 cm/s) pH 5.0/6.3/7.4 (AID-1557) | Plasma Protein Binding (% Bound) (AID-1617) | Plasma Stability (%Remaining) Human/Mouse (AID-1591/AID-1592) | Microsomal Stability (%Remaining) Human/Mouse (AID-1555/pending) | |
|---|---|---|---|---|---|---|
| Human (10μM/1μM) | Mouse (10μM/1μM) | |||||
| CID-665093 MLS-0014097 | 8.4/8.4/18.0 | 90/98/58 | 98.6/98.7 | 87.9/89.5 | 100.2/83.7 | 47.0/41.8 |
| CID-2102207 MLS-0107074 | 0.3/0.3/0.9 | 98/91/75* | 99.9/99.1 | 99.7/99.3 | 82.4/77.5 | 17.0/1.8 |
CID-665093 (MLS-0014097) exhibits modest solubility in aqueous buffer and poor cell permeability. On the other hand, since PLAP is an extracellular enzyme, poor permeability could provide an added advantage to the probe, helping to ensure selectivity for its biological function. The compound is most soluble in aqueous buffer at pH 7.4, indicating that the compound will be sufficiently soluble for use in cellular assays conducted under normal culture conditions. CID-665093 (MLS-0014097) is extensively bound to plasma proteins in human plasma. In mouse plasma, the percent compound bound is slightly less than in human plasma. The amount of free compound available to bind the target (PLAP) is therefore greater in mice suggesting that the efficacy of this compound might be greater in mice than in humans. The compound also exhibits robust metabolic stability when exposed to both mouse and human microsome preparations.
CID-2102207 (MLS-0107074) exhibits poor solubility in aqueous buffer at all pH levels tested. In order to assess the cellular permeability, it was required that a cosolvent (20% ACN) be used in the assay to facilitate solubility. Despite the use of ACN, cellular permeability remained poor. The compound is extensively bound to plasma proteins in human and mouse plasma, and exhibits moderate stability in human and mouse plasma. CID-2102207 (MLS-0107074) was significantly metabolized in human and mouse hepatic microsomes. This suggests that the compound will be subjected to extensive first pass metabolism, limiting the use of the compound in vivo. Taken together these attributes indicate that this probe is best suited for biochemical experiments, but may also be used in cell based experiments when formulated with an appropriate co-solvent.
j. Properties Computed from Structure
| Property | Value |
|---|---|
| Molecular Weight | 397.44756 [g/mol] |
| Molecular Formula | C20H19N3O4S |
| XLogP3-AA | 3.3 |
| H-Bond Donor | 2 |
| H-Bond Acceptor | 6 |
| Rotatable Bond Count | 8 |
| Tautomer Count | 22 |
| Exact Mass | 397.109627 |
| MonoIsotopic Mass | 397.109627 |
| Topological Polar Surface Area | 123 |
| Heavy Atom Count | 28 |
| Formal Charge | 0 |
| Complexity | 538 |
| Isotope Atom Count | 0 |
| Defined Atom StereoCenter Count | 0 |
| Undefined Atom StereoCenter Count | 0 |
| Defined Bond StereoCenter Count | 0 |
| Undefined Bond StereoCenter Count | 0 |
| Covalently-Bonded Unit Count | 1 |
4. Appendices
a. Comparative data on (1) probe, (2) similar compound structures (establishing SAR) and (3) prior probes
Table 5All Purchased Analogs
| Structure | Cmpd ID | IC50(µM) | SID | CID | Vendor | VendorID | |
|---|---|---|---|---|---|---|---|
| PLAP | TNAP | ||||||
 | MLS-0014097 | 2.6 | >100 | 56373725 | 665093 | InterBio-Screening | STOCK4S-90308 |
 | MLS-0390850 | 13.6 | >100 | 57287596 | 1400698 | InterBio-Screening | STOCK4S-91062 |
 | MLS-0390843 | 3.97 | >100 | 57287589 | 1409759 | InterBio-Screening | STOCK4S-87186 |
 | MLS-0390851 | 6.02 | >100 | 57287597 | 1400246 | InterBio-Screening | STOCK4S-92693 |
 | MLS-0390858 | 9.4 | >100 | 57287605 | 1394536 | InterBio-Screening | STOCK5S-03688 |
 | MLS-0390855 | >100 | >100 | 57287601 | 1409691 | InterBio-Screening | STOCK4S-97188 |
 | MLS-0390857 | 7.45 | >100 | 57287604 | 1398345 | InterBio-Screening | STOCK4S-99075 |
 | MLS-0390849 | 11.8 | >100 | 57287595 | 1400269 | InterBio-Screening | STOCK4S-90841 |
 | MLS-0390841 | 2.7 | 12.1 | 57287587 | 1416608 | InterBio-Screening | STOCK4S-84785 |
 | MLS-0297802 | 2.92 | >100 | 57287584 | 1400947 | InterBio-Screening | STOCK4S-15744 |
 | MLS-0108835 | 2.53/ | 40.3 | 56373724 | 1399636 | InterBio-Screening | STOCK4S-90782 |
 | MLS-0101351 | 11.8 | >100 | 57287602 | 3878415 | InterBio-Screening | STOCK4S-97273 |
 | MLS-0390844 | 8.76 | >100 | 57287590 | 1398070 | InterBio-Screening | STOCK4S-88147 |
 | MLS-0390842 | 8.31 | >100 | 57287588 | 1402438 | InterBio-Screening | STOCK4S-86311 |
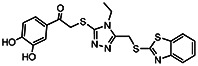 | MLS-0107074 | 3.87 | >100 | 57309171 | 2102207 | BCCG | PMK-14-59 |
 | MLS-0315681 | 2.79 | 93.4 | 56373578 | 838842 | Chem-Bridge | 5672011 |
 | MLS-0390856 | 11.7 | >100 | 57287603 | 1400150 | InterBio-Screening | STOCK4S-97686 |
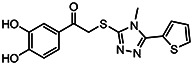 | MLS-0297827 | 4.9 | >100 | 57287583 | 3735808 | InterBio-Screening | STOCK3S-98364 |
 | MLS-0315688 | 7.29 | 2.78 | 56373639 | 1314069 | Chem-Bridge | 7587885 |
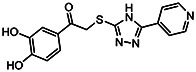 | MLS-0315684 | 6.95 | 6.53 | 56373634 | 715398 | Chem-Bridge | 6593203 |
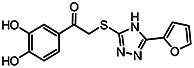 | MLS-0315686 | 12.2 | 21.7 | 56373636 | 715363 | Chem-Bridge | 6617586 |
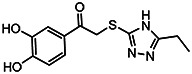 | MLS-0315685 | 1.78 | 10.9 | 56373635 | 715330 | Chem-Bridge | 6607811 |
 | MLS-0390847 | >100 | >100 | 57287593 | 1400320 | InterBio-Screening | STOCK4S-90030 |
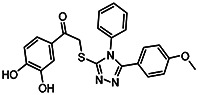 | MLS-0390853 | 7.22 | >100 | 57287599 | 1400627 | InterBio-Screening | STOCK4S-94912 |
 | MLS-0390852 | 8.73 | >100 | 57287598 | 1399934 | InterBio-Screening | STOCK4S-94867 |
 | MLS-0390854 | 5.59 | >100 | 57287600 | 1401217 | InterBio-Screening | STOCK4S-95328 |
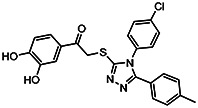 | MLS-0390846 | 13 | >100 | 57287592 | 1383304 | InterBio-Screening | STOCK4S-89768 |
 | MLS-0390839 | 25.0 | >100 | 57287585 | 1973575 | InterBio-Screening | STOCK4S-16633 |
 | MLS-0390840 | 6.63 | 40.7 | 57287586 | 1984977 | InterBio-Screening | STOCK4S-20993 |
 | MLS-0390848 | 4.94 | 19.2 | 57287594 | 4283152 | InterBio-Screening | STOCK4S-90562 |
 | MLS-0315690 | 1.58 | 8.58 | 56373641 | 946537 | Chem-Bridge | 7746693 |
 | MLS-0238024 | 1.89 | 5.98 | 56373642 | 713665 | Chem-Bridge | 7764114 |
 | MLS-0008112 | 1.42 | 5.93 | 56373572 | 1515366 | Chem-Bridge | 7764254 |
 | MLS-0255212 | 1.76 | 3.48 | 56373643 | 2010274 | Chem-Bridge | 7775407 |
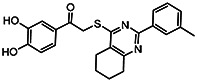 | MLS-0315689 | >100 | 78.5 | 56373640 | 1305489 | Chem-Bridge | 7642884 |
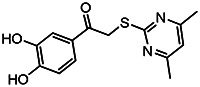 | MLS-0315687 | 1.85 | 7.19 | 56373637 | 715454 | Chem-Bridge | 6622859 |
 | MLS-0315680 | 1.56 | 6.71 | 56373577 | 720372 | Chem-Bridge | 5484218 |
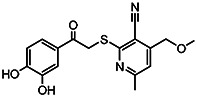 | MLS-0051931 | 2.16 | 4.82 | 56373536 | 586278 | ChemDiv | 3294-0014 |
 | MLS-0315683 | 8.77 | >100 | 56373633 | 5350662 | Chem-Bridge | 6588132 |
 | MLS-0315887 | 4.79 | 5.44 | 56373703 | 4206177 | Specs | AI-204/31718048 |
 | MLS-0315886 | 3.06 | 4.15 | 56373702 | 4450383 | Specs | AI-204/31685046 |
 | MLS-0315885 | 3.42 | 3.03 | 56373701 | 4066568 | Specs | AI-204/31680049 |
 | MLS-0315884 | >100 | >100 | 56373700 | 996625 | Specs | AG-690/40754100 |
 | MLS-0315888 | >100 | >100 | 56373704 | 698547 | Specs | AM-807/12427007 |
 | MLS-0315883 | >100 | >100 | 56373696 | 1223257 | Specs | AE-848/36959517 |
 | MLS-0315682 | >100 | 3.71 | 56373579 | 2877377 | Chem-Bridge | 5933089 |
 | MLS-0315889 | >100 | >100 | 56373705 | 715329 | Specs | AP-048/15613011 |
 | MLS-0080017 | >100 | >50 | 56373623 | 881297 | Chem-Bridge | 6385545 |
 | MLS-0315678 | 39.3 | >100 | 56373922 | 2830496 | Chem-Bridge | 5147768 |
 | MLS-0315679 | 18.8 | >100 | 56373923 | 2841226 | Chem-Bridge | 5317936 |
 | MLS-0390862 | >100 | >100 | 57287609 | 16415295 | InterBio-Screening | STOCK6S-42849 |
 | MLS-0390859 | 0.374 | 3.14 | 57287606 | 5922288 | InterBio-Screening | STOCK1S-39868 |
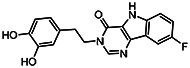 | MLS-0390860 | 0.93 | 0.25 | 57287607 | 932151 | InterBio-Screening | STOCK2S-62329 |
 | MLS-0390861 | 15 | 7.38 | 57287608 | 16000021 | InterBio-Screening | STOCK4S-15750 |
 | MLS-0077534 | 2.95 | 29.2 | 56373574 | 2940433 | Chem-Bridge | 7471251 |
 | MLS-0068603 | >100 | >100 | 56373638 | 805519 | Chem-Bridge | 6680720 |
 | MLS-0073729 | 8.43 | >100 | 56373913 | 9551841 | InterBio-Screening | STOCK2S-13283 |
b. Comparative data showing probe specificity for target
Table 4SAR and Selectivity
| Compound ID | Assay Format | Assay Target | No of Runs | IC50 Value (μM) |
|---|---|---|---|---|
| MLS-0014097 | Luminescent | TNAP | 4 | >100 |
| MLS-0014097 | Luminescent | PLAP | ~ 6 | 2.34 |
| MLS-0014097 | Luminescent | IAP | 1 | 20.7 |
| MLS-0014097 | Colorimetric | GAPDH | 2 | >100 |
| MLS-0014097 | FP | BCL-b/TR3 | 1 | >100 |
| MLS-0014097 | FP | Bfl-1 | 1 | >100 |
5. Bibliography
- 1.
- Hoylaerts MF, Manes T, Millán JL. Molecular mechanism of uncompetitive inhibition of human placental and germ-cell alkaline phosphatase. Biochem J. 1992;286(Pt 1):23–30. [PMC free article: PMC1133013] [PubMed: 1520273]
- 2.
- Wennberg C, Kozlenkov A, Di Mauro S, Fröhlander N, Beckman L, Hoylaerts MF, Millán JL. Structure, genomic DNA typing, and kinetic characterization of the D allozyme of placental alkaline phosphatase (PLAP/ALPP). Human Mutation. 2002;19(3):258–267. [PubMed: 11857742]
- 3.
- Wahren B 1, Hinkula J, Stigbrand T, Jeppsson A, Andersson L, Esposti PL, Edsmyr F, Millán JL. Phenotypes of placental-type alkaline phosphatase in seminoma sera. Int J of Cancer. 2006;37(4):595–600. [PubMed: 3957465]
- 4.
- Fishman WH. Clinical and biological significance of an isozyme tumor marker—PLAP. Clin Biochem. 1987;20:387–92. [PubMed: 3325192]
- PMCPubMed Central citations
- PubChem BioAssay for Chemical ProbePubChem BioAssay records reporting screening data for the development of the chemical probe(s) described in this book chapter
- PubChem SubstanceRelated PubChem Substances
- PubMedLinks to PubMed
- Review Placental Alkaline Phosphatase (PLAP) Luminescent HTS assay - Probe 2.[Probe Reports from the NIH Mol...]Review Placental Alkaline Phosphatase (PLAP) Luminescent HTS assay - Probe 2.Lanier M, Cashman J, Sergienko E, Garcia X, Brown B, Rascon J, Narisawa S, Simao AM, Millán JL, Stonich D, et al. Probe Reports from the NIH Molecular Libraries Program. 2010
- Review A Selective Murine Intestinal Alkaline Phosphatase (muIAP) Inhibitor.[Probe Reports from the NIH Mol...]Review A Selective Murine Intestinal Alkaline Phosphatase (muIAP) Inhibitor.Ganji S, Zou J, Brown B, Bobkova EV, Ardecky R, Rosenstein C, Pass I, Dahl R, Vasile S, Sergienko E, et al. Probe Reports from the NIH Molecular Libraries Program. 2010
- Heterogeneity in alkaline phosphatase isozyme expression in human testicular germ cell tumours: An enzyme-/immunohistochemical and molecular analysis.[J Pathol. 1999]Heterogeneity in alkaline phosphatase isozyme expression in human testicular germ cell tumours: An enzyme-/immunohistochemical and molecular analysis.Roelofs H, Manes T, Janszen T, Millán JL, Oosterhuis JW, Looijenga LH. J Pathol. 1999 Oct; 189(2):236-44.
- Levels of alkaline phosphatase isozymes in human seminoma tissue.[Cancer Res. 1987]Levels of alkaline phosphatase isozymes in human seminoma tissue.Hirano K, Domar UM, Yamamoto H, Brehmer-Andersson EE, Wahren BE, Stigbrand TI. Cancer Res. 1987 May 15; 47(10):2543-6.
- Modulators of intestinal alkaline phosphatase.[Methods Mol Biol. 2013]Modulators of intestinal alkaline phosphatase.Bobkova EV, Kiffer-Moreira T, Sergienko EA. Methods Mol Biol. 2013; 1053:135-44.
- Placental Alkaline Phosphatase (PLAP) Luminescent HTS assay - Probe 1 - Probe Re...Placental Alkaline Phosphatase (PLAP) Luminescent HTS assay - Probe 1 - Probe Reports from the NIH Molecular Libraries Program
Your browsing activity is empty.
Activity recording is turned off.
See more...