NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
Chagas disease is a tropical disease caused by the parasite Trypanosoma cruzi (T. cruzi), which is endemic to Central and South America. Approximately 13 million people are infected with the parasite, and 25–30% of infected patients suffer from irreversible damage to the heart and digestive tract resulting in disability and death within 20 years of infection. Few treatments are available with limited effectiveness only against the early (acute) stages of disease, significant toxicity, and widespread drug resistance. We report the outcome of a high-throughput chemical library screen to identify novel, nontoxic, small-molecule inhibitors of T. cruzi, which will aid the development of more potent and selective therapies for both the acute and chronic stages of Chagas disease. Of the 303,224-screened compounds, 35 compounds were chosen based on their selectivity, potency, and chemical tractability. Of those, 27 dry powder-validated compounds were retested in the primary screen, secondary assays, and an orthogonal screen. Three scaffolds were prioritized to identify potential probes. One of these compounds (CID3238551/ML158) displayed greater than 100-fold selective inhibition of T. cruzi confirmed in immunofluorescent imaging assays and is inactive against host cells at the highest tested dose. This new probe should be very useful in future cell-based investigations and in vivo studies of T. cruzi inhibition.
Assigned Assay Grant #: 1R03MH085673-01
Screening Center Name and PI: Broad Institute Probe Development Center, Stuart Schreiber
Chemistry Center Name and PI: Broad Institute Probe Development Center, Stuart Schreiber
Assay Submitter and Institution: Ana Rodriguez, New York University
PubChem Summary Bioassay Identifier (AID): 1885
Probe Structure and Characteristics
Compound Summary in PubChem
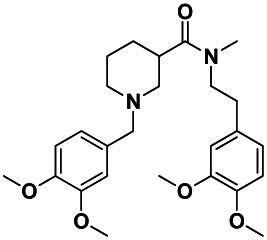
| IUPAC Chemical Name | N-[2-(3,4-dimethoxyphenyl)ethyl]-1-[(3,4- dimethoxyphenyl)methyl]-N-methylpiperidine-3- carboxamide |
| PubChem CID | 3238551 |
| Molecular Weight | 456.57444 g/mol |
| Molecular Formula | C26H36N2O5 |
| XLogP | 3.6 |
| H-Bond Donor | 0 |
| H-Bond Acceptor | 6 |
| Rotatable Bond Count | 10 |
| Exact Mass | 456.262422 |
| Topological Polar Surface Area | 60.5 |
| Solubility (PBS, μM) | >500 |
| CID/MLS No. | Target Name | IC50/EC50 (nM) [SID, AID] | Anti-target | IC50/EC50 (μM) [SID, AID] | Fold Selective | Secondary Assay(s) [SID, AID] |
|---|---|---|---|---|---|---|
| 3238551/ML158 | Trypansoma cruzi replication | 109 [SID 87219036, AID 2630] | NIH/3T3 Cell Toxicity | Inactive [SID 87219036, AID 2586] | 109 nM vs inactive | Immunofluorescence Static [SID 87219036, AID 2630] |
Recommendations for scientific use of the probe
The goal of this project is to identify a small-molecule inhibitor of the protist Trypanosoma cruzi (T. cruzi), the causative parasite of Chagas disease. Although a few treatments for Chagas disease are available, these treatments are limited because they are only effective against the early (acute) stages of disease and have significant toxicity to the patient (1,2,3). Novel antitrypanosomal agents will aid in developing a drug treatment for both the acute and chronic stages of Chagas disease. This probe (CID3238551/ML158) is expected to inhibit the replication of T. cruzi without having effects on the viability of the mammalian host cell.
The probe (CID3238551/ML158) described in this report inhibits the replication of T. cruzi with an IC50 of 109 nM and is inactive toward mammalian cells, NIH/3T3 up to concentrations of 19,500 nM. In immunofluorescent imaging assays, the compound inhibited T. cruzi amastigote replication within host cells. Since amastigote morphology was maintained, this probe (CID3238551/MLS002725691) is categorized as a trypanostatic (static) compound.
This probe (CID3238551/ML158) provides a valuable tool that will allow microbiologists investigating Chagas disease to identify new targets in T. cruzi, which may potentially lead to better drugs. Furthermore, the Assay Provider, Dr. Ana Rodriguez and her colleagues, will continue their own studies with this probe (CID3238551/ML158) in a mouse model of Chagas disease.
1. Introduction
Scientific Rationale
Chagas disease is a tropical parasitic disease caused by the parasite Trypanosoma cruzi (T. cruzi). Approximately 13 million people are infected with the parasite, which is endemic to Central and South America and results in 14,000 deaths per year (2, 3). Twenty-five to thirty percent of infected patients suffer from irreversible damage to the heart and digestive tract resulting in a high incidence of disability and death within 20 years of infection (1, 2). Chagas disease is the leading cause of heart failure in the region.
In Figure 1, the drugs currently used to treat Chagas disease are: benznidazole A (Rochagan®, Radanil® Hoffman Roche) and nifurtimox B (Lampit®, Bayer), whose anti-T. cruzi activities were discovered more than 30 years ago. Both of these drugs are effective only in the acute phase of infection, rather than the key chronic stage. They often do not completely eliminate the parasite despite long-term administration, while exhibiting unacceptable side effects (such as nausea, vomiting, weight loss, neurological effects, and signs of testicular and ovarian injury). Both treatments are actually nonspecific for T. cruzi; in addition, resistance to these drugs is becoming more widespread. Thus, there is a clear need for more potent and selective therapies.

Figure 1
Current Treatments for Chagas Disease. Benznidazole (Rochagen®, Radanil®, Hoffman-La Roche) (A); Nifurtimox (Lampit®, Bayer) (B)
Several recent reports have revealed promising new lead compounds that target T. cruzi, including inhibitors of the cysteine protease cruzain and inhibitors of sterol demethylase (4). However, while the protease inhibitors (see Figures 2A, 2B, and 2C) exhibit excellent activity in biochemical assays (0.079 μM to 2 μM), they are inactive or demonstrate high micromolar activity in parasite inhibition in cell culture (1 μM to inactive) (4, 5, 6). Thus, there is a continued unmet need to identify scaffolds that are potent in cell culture against T. cruzi and nontoxic to the mammalian host cell. Our approach overcomes this problem of lack of cellular activity by employing a phenotypic screen measuring parasite replication in a host cell. Counterscreening in the mammalian host filters out toxic compounds.
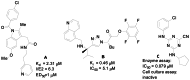
Figure 2
Recent Inhibitors of T. cruzi Cysteine Proteases.
There are currently four compounds in clinical trials for Chagas disease (see Figure 3). Benznidazole (see Figure 1A) initially developed for treatment of acute Chagas disease is in Phase III trials to expand its use to include the treatment of chronic Chagas cardiomyopathy. Three antifungal drugs from Merck (see Figure 3A), Takeda (see Figure 3B), and Eisai (structure not disclosed) are in Phase I or II trials for expanding their use to include treatment of Chagas disease. Two heart arrhythmia treatments (see Figure 3C and Figure 3D) are the most frequent treatments for the chronic stage heart issues associated with Chagas disease and have shown anti-parasitic activity in some patients. The only novel compound currently in the pipeline is a peptide mimic from the Sandler Center for Drug Discovery (see Figure 3E) which is in preparation for a Phase I safety trial. Since all of these compounds are nonspecific for T. cruzi, the side effects may be problematic as is the case for benzidazole in its current use. Therefore, a selective, nontoxic inhibitor of T. cruzi replication could be of significant use to the scientific community.

Figure 3
Compounds in Clinical Trials for Chagas Disease.
This project aims to identify novel antitrypanosomal agents for the treatment of Chagas disease. One key design of the project is to target the parasites co-cultured with host cells and to demonstrate little to no toxicity to the host cell. This screening campaign has identified a probe, N-[2-(3,4-dimethoxyphenyl)ethyl]-1-[(3,4-dimethoxyphenyl)methyl]-N-methylpiperidine-3-carboxamide (CID3238551/ML158), which has an IC50 of 109 nM and has no toxicity towards NIH/3T3 cells at the highest concentration tested (i.e., 19.5 μM).
2. Materials and Methods
The methods in this section were either performed as described in Bettiol et al. (i.e., immunofluorescence) (7) or modified for high throughput screening (co-culture and host cell toxicity).
Materials and Reagents
T175 culture flasks with vented caps were obtained from BD Falcon, and hyperflasks were obtained from Corning (Corning, NY; Catalog no.10024). Disposable sterile filter units (500 ml or 1 L; pore size, 0.20 μm were obtained from Nalgene (Catalog no. 566-0020). Dulbecco’s modified Eagle’s medium (DMEM) with Phenol Red, high glucose, with L-glutamine and sodium pyruvate was obtained from Cellgro (Mediatech Inc, Manassas, VA; Catalog no. 10-013-CM). Penicillin-streptomycin-L-glutamine (PSG, Catalog no. 10378-016), FBS-heat inactivated fetal bovine serum (FBS, Catalog no.16140-089), and 0.25% Trypsin-EDTA 1X (Catalog no. 25200-072) were purchased from Gibco-Invitrogen. Sterile horse serum, from donor herd (if appearance of epimastigotes) was obtained from Sigma (Catalog no. H1270). Sterile, Ca++/Mg++-free Phosphate Buffered Saline (PBS) 1X was prepared in house.
Nonidet P-40 (NP40, now called Igepal CA 360) was obtained from Fluka (Sigma-Aldrich, St. Louis, MO; Catalog no. 56741) and Gal-Screen® Buffer B was obtained from AB Biosciences (Allston, MA; Catalog no.T1031).
Cell Lines
- The following cell lines were used in this study: LLC-MK2 cells (rhesus monkey kidney epithelial cell line) and NIH/3T3 cells (mouse embryonic fibroblastic cell line) were initially obtained from the Assay Provider, then from ATCC. T. cruzi expressing β-galactosidase (T. cruzi -β-gal: Tulahuen strain, clone C4; was obtained from the Assay Provider with derivation as described in Buckner et al. (8).
- Alexa Fluor 488 goat anti-rabbit IgG secondary antibody was from Molecular Probes®, Invitrogen (Carlsbad, CA).
- Polyclonal rabbit anti-T. cruzi was a gift from Dr. B. Burleigh, Harvard School of Public Health, Boston, MA).
2.1. Assays
A summary listing of completed assays and corresponding PubChem AID numbers is provided in Appendix A (Table A1). Refer to Appendix B for the detailed assay protocols.
2.1.1. T. cruzi Inhibition Assay (AID Nos.1885, 2044, 2294, 2630)
For cell propagation: 90% DMEM, Phenol Red, 10% FBS, and 1% PSG were mixed and filtered through a 0.2 micron membrane. The cells were kept at 4°C, and then warmed up to 37°C in a water bath before use.
For T. cruzi culture and assays: 98% DMEM, Phenol Red, 2% FBS, and 1% PSG were mixed and filtered through a 0.2 micron membrane. The cells were kept at 4°C, and then warmed up to 37°C in a water bath before use.
Solutions: Gal-Screen + 0.05% NP40. Using a Gal-Screen base kit, Buffer B (Catalog no. T2361) was mixed with 1:25 substrate (Catalog no. T2359) with a 1:400 dilution of 20% NP40.
NIH/3T3 Cell Culture. NIH/3T3 cells were cultivated in DMEM supplemented with 10% FBS and 1% PSG in T175 in 50 ml total of medium.
LLC-MK2 Cell Culture. LLC-MK2 cells were cultivated in DMEM supplemented with 10% FBS and 1% PSG in T175 flasks in 50 ml total of medium. Cells were usually passaged twice a week at 1:4 to 1:8 ratios.
Parasite Culture: Tcruzi β-gal (Tc). T. cruzi -β-gal were cultivated in DMEM supplemented with 2% FBS and 1% PSG in T175 flasks with vented caps (important to avoid spills!) in 50 ml total of medium.
2.1.2. Growth Inhibition Assay for HTS (384-well plates)
The medium was warmed up with 2% FBS/DMEM. The parasites were harvested in 50-ml tubes, and spun for 10 minutes at 2200 rpm. Approximately 15 ml of media was aspirated, and the samples were incubated for 3–5 hours. The NIH/3T3 cells were trypsinized (refer to cell culture protocol). When the NIH/3T3 cells were detached, the cells were harvested in DMEM, 2% FBS, and 1% PSG, then counted using the Nexcelom Cellometer. The cells were diluted to 166,667 cells/ml, and then added to a flask and plated 5,000 cells/30 μL per well using a standard cassette multiwell drop Combi. The cells were incubated for 3 hours, and then T. cruzi cells were counted, diluted to 0.250 million cells/ml, and transferred to a 2-liter flask. Then, 50 nL compounds/DMSO were pinned to each well with NIH/3T3 cells. Next, 20 μL/well of parasites (5000 T. cruzi) were added with a standard cassette multiwell drop Combi on slow speed, and incubated for 4 days (or a minimum of 90 hours). Gal-Screen was prepared with 0.05% NP40, 30 μL per well were dispensed in a 384-well plate, incubated for 60 minutes, and the luminescence was read using Envision (Perkin-Elmer) at 0.1 sec/well.
2.1.3. Cell Toxicity Assay: NIH/3T3 Cells (AID Nos. 2010, 2586, 493247)
For the cell toxicity assay with NIH/3T3 cells, the same materials as for T. cruzi co-culture assay were used. NIH/3T3 cells were cultivated in DMEM supplemented with 10% FBS and 1% PSG in T175 in 50 ml total of medium.
2.1.4. Intracellular T. cruzi Immunofluorescence Assay (AID No. 2632)
Fifty thousand NIH/3T3 cells were seeded on sterile glass coverslips in 12-well plates and allowed to adhere overnight. Five million T. cruzi parasites were added (Mechanism of inhibition 100:1) and allowed to infect for 2 hours in DMEM+2% FBS and PSG. Parasites were rinsed out 3X with PBS, and compounds were added at 10X their IC50 (as determined in AID 2044 and AID 2294). Infected cells were further incubated for 4 days and fixed for 15 minutes with 4% paraformaldehyde.
Fixed cells on coverslips were rinsed with PBS and permeabilized for 15 minutes in PBS with 0.1% Triton X-100. After blocking for 20 minutes in PBS with 10% goat serum, 1% bovine serum albumin (BSA), 100 mM glycine, and 0.05% sodium azide, cells were incubated for 1 hour at room temperature with a polyclonal rabbit anti-T. cruzi at 1:2000 dilution. After rinsing, an Alexa Fluor 488 goat anti-rabbit IgG secondary antibody was added for 1 hour at a 1:800 dilution. DNA was stained with DAPI, and coverslips were mounted with anti-fade mounting media. Images were taken using an inverted Olympus IX70 microscope with a 60X oil objective.
2.2. Probe Chemical Characterization
The probe (CID3238551/ML158) 1 was purchased from ChemDiv (ID 5979-0184); however, the probe could be synthesized starting from nipecotic acid in three steps as shown in Scheme 1. The above route was used to synthesize several analogs. Amide coupling of nipecotic acid 11 with 3,4-dimethoxyphenethyl amine would result in the amide 12. Reductive amination of 12 with 3,4-dimethoxybenzaldehyde would provide probe 1 (CID3238551/ML158). The probe (CID3238551/ML158) was determined to have a solubility of >500 μM in PBS at room temperature.

Scheme 1
Synthesis of Probe (CID3238551/ML158).
The probe (CID3238551/ML158) and five analogs were submitted to the SMR collection (MLS002725691, MLS002725685, MLS002725682, MLS002725692, MLS002725694).
The 1H NMR (300 MHz, DMSO-d6) spectra and LC-MS chromatograms of the probe (CID3238551/ML158) and analogs (CID16187238, CID16191661, CID44629407, CID44629409, CID44629410, CID44629413, and CID 16190091) are provided in Appendix C.
2.3. Probe Preparation
Not applicable. The probe (CID3238551/ML158) was purchased from ChemDiv (ID 5979-0184).
3. Results
Probe Attributes
- Inhibits extracellular parasite invasion into host cell or inhibits intracellular parasitic replication.
- Non-toxic to host cells (100-fold selectivity towards parasite vs. host cells).
3.1. Summary of Screening Results
Figure 4 displays the critical path for probe development.
A high-throughput screen of 303,224 compounds (AID 1885) was performed in duplicate in the recombinant Tulahuen strain of T. cruzi stably expressing beta-galactosidase reporter co-cultured with host cell, mouse fibroblast NIH/3T3 (8). The signal was normalized to neutral (DMSO) controls, and a 55% inhibition cutoff at a screening concentration of 3.75 μM average was used to define a hit. Next, 4,394 hits were identified as inhibitors of T. cruzi replication when co-cultured with host cells and, of these, 4,063 were available as cherry picks. The cherry-picked compounds were retested in dose in the primary assay using co-cultured T. cruzi strain with host cells to confirm their inhibitory activity (AIDs 2044, 2294). From the 4,063 compounds retested at dose concentrations, 4,016 compounds (99%) re-confirmed in this assay. In parallel, these 4,063 compounds were included in toxicity assays against host cell NIH/3T3 (AID 2010) to determine if these compounds were cytotoxic to mammalian cells and, thus, false positives. This secondary screen identified 1,011 compounds that reduced viability of NIH/3T3 cells; therefore, these compounds were excluded as viable hits.
After completing retests and a secondary assay at dose from DMSO stocks, 3,005 compounds were clustered into groups with 70% similarity. The 25 clusters with at least five members were given to a team of chemists; 35 representative compounds, at least one from each cluster, were prioritized based on molecular weight, presumed solubility, and lack of toxic or reactive functionality. Of the 35 compounds selected, 27 authentic dry powders were obtained. From this set of dry powder compounds, a piperidine series was prioritized. Additional analogs were ordered and tested in the primary and secondary toxicity assay. The results are reported in Table 1, Table 2, and Table 3. Also, these compounds were tested in an additional immunofluorescence assay to determine the mode of action of the compounds (AID 2632).
Table 1SAR Analysis of Dimethoxy-phenethyl Amide Series
| SAR Analysis for T. cruzi |
 | Potency (μM) mean ± S.E.M. (n=replicates) | Target to Antitarget Fold Selectivity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Entry | CID | SID | Broad No. | * | R | n | T. cruzi inhibition | n | Toxicity | |
| 1 | 3238551 | 87219048 | BRD-A98248982 | P |
 | 3 | 0.109 ± 6.5 | 3 | inactive | >500 |
| Solubility: 105.4 μM | Purity: 94% | |||||||||
| 2 | 3377343 | 99351065 | BRD-A54545770 | P |
 | 3 | 1.4 ± 0.8 | 3 | 47.4 | 34 |
| Solubility: ND | Purity: >95% | |||||||||
| 3 | 46897906 | 99351063 | BRD-A72455176 | P |
 | 3 | 4.33 ± 1.76 | 3 | 64.7 ±21.5 | 15 |
| Solubility: ND | Purity: >95% | |||||||||
ND= Not determined;
- *
P = Purchased
Table 2SAR Analysis of the Benzyl Amide Series
| SAR Analysis for T. cruzi |
 | Potency (μM) mean ± S.E.M. (n=replicates) | Target to Antitarget Fold Selectivity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Entry | CID | SID | Broad No. | * | R | n | T. cruzi inhibition | n | Toxicity | |
| 1 | 46912157 | 99376496 | BRD-A84438227 | S |
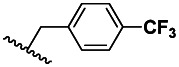 | 3 | 0.008±0.005 | 3 | 61.6 ±29.5 | >500 |
| Solubility: ND | Purity: 81% | |||||||||
| 2 | 3500831 | 87219049 | BRD-A83402799 | P |
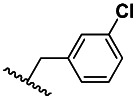 | 3 | 0.570±0.247 | 3 | 70.3 ±67.5 | 123 |
| Solubility: >500 μM | Purity: >95% | |||||||||
| 3 | 16470923 | 99376495 | BRD-A30944476 | S |
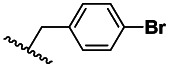 | 3 | 3.01 ±1.12 | 3 | 74.3 ±18.5 | 25 |
| Solubility: ND | Purity: >95% | |||||||||
| 4 | 46912177 | 99376497 | BRD-A51613646 | S |
 | 3 | 7.63 ± 5.05 | 3 | 51.0 ±14.9 | 6.7 |
| Solubility: ND | Purity: >95% | |||||||||
| 5 | 3237704 | 87556793 | BRD-A74871599 | P |
 | 3 | 10.8 ±4.45 | 3 | 77.3 ±58.4 | 7.2 |
| Solubility: ND | Purity: >95% | |||||||||
| 6 | 87556794 | 2930928 | BRD-A16581344 | P |
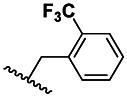 | 3 | 15.9 ±4.48 | 3 | 63.3 ±37.6 | 7.2 |
| Solubility: >500 μM | Purity: >95% | |||||||||
ND= Not determined;
- *
P = Purchased; S = Synthesized
Table 3SAR Analysis for T. cruzi
| SAR Analysis for T. cruzi |
 | Potency (μM) mean ± S.E.M. (n=replicates) | Target to Antitarget Fold Selectivity | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Entry | CID | SID | Broad No. | * | R | R′ | n | T. cruzi inhibition | n | Toxicity | |
| 1 | 46897916 | 99351067 | BRD-A04542699 | P |
 |
 | 3 | 0.763 ±0.0413 | 3 | 52.7 ±19.9 | 69 |
| Solubility: ND | Purity: 91% | ||||||||||
| 2 | 4207918 | 87556801 | BRD-A99518825 | P |
 |
 | 3 | 1.01 ±0.655 | 3 | inactive | >500 |
| Solubility: >500 μM | Purity: 93% | ||||||||||
| 3 | 99351072 | 3422467 | BRD-A36791463 | P |
 |
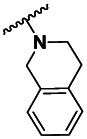 | 3 | 3.54 ±2.12 | 3 | 41.7 ±6.96 | 12 |
| Solubility: 388.9 μM | Purity: 88% | ||||||||||
| 4 | 87556795 | 2838441 | BRD-A98050686 | S |
 |
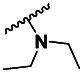 | 3 | 9.91 ±3.67 | 3 | 164 ±53.1 | 17 |
| Solubility: >500 μM | Purity: 85% | ||||||||||
ND= Not determined;
- *
P = Purchased; S = Synthesized
Intracellular T. cruzi was treated with the probe (CID3238551/ML158). The compound inhibited replication of T. cruzi within the mammalian host cell but did not lyse T. cruzi. Therefore, the compound was classified as a ‘static’ inhibitor. Related analogs were also tested (AIDs 2630, 2586, 2632).
3.2. Dose Response Curves for Probe
Figure 5 displays dose response curves for the probe.
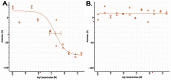
Figure 5
Dose-dependent Activities of Probe (CID3238551/ML158) in Target and Counterscreen Assays. Primary screen-from dry powders (IC50 109 nM) (AID 2630) (A); NIH3T3 (inactive) Toxicity (AID 2586) (B).
The probe (CID3238551/ML158) met the defined probe criteria by displaying greater than 100-fold selective inhibition of T. cruzi versus NIH/3T3 (109 nM versus inactive). Furthermore, the inhibition of T. cruzi replication was confirmed in immunofluorescent imaging assays (AID 2632). The compound inhibited replication as compared to DMSO control conditions, and the compound did not lyse the cells. The probe (CID3238551/ML158) has acceptable solubility and stability in aqueous conditions.
3.3. Scaffold/Moiety Chemical Liabilities
A search of PubChem for the probe (CID3238551/ML158) revealed that the probe has been tested in 359 BioAssays and was confirmed as active only in our assays. Therefore, the compound is not promiscuous. The compound has a drug-like structure with no obvious chemical liabilities that would be a concern.
The stability of the probe (CID3238551/ML158) in PBS was monitored over 48 hours, and the data is presented in Figure 6. After 24 hours, 100% of the compound was remaining. The amount of compound decreased to 84.2% between 24 to 48 hours. Recovery with acetonitrile was done at each time point to account for any precipitation and showed that no precipitation occurred.

Figure 6
PBS Stability of the Probe (CID3238551/ML158).
3.4. SAR Tables
Screening results for the primary assay are described in the above tables. The most potent IC50 of inhibition without toxicity was measured with compound 1 (CID3238551/ML158), which has been designated as the probe for this series. As can be seen in Table 1, substitutions at the 3 and 4 positions of the benzyl amine substituent seem to be tolerable, while substitution at the 2 position results in a significant decrease in activity.
The substitution preference is reinforced by the results shown in Table 2. The 4-trifluoromethyl substituted compound (CID46912157) was the most potent compound at 8 nM; however, it showed a significant increase in toxicity over the probe. The 2-trifluoromethyl substituted compound (CID87556794) was one of the least potent compounds tested at 15.9 μM; however, it had similar toxicity.
Replacement of the amide with other amides resulted in a decrease in activity; however, tying the amide back (CID46897916) resulted in similar activity (0.765 μM) when compared to the benzyl substituted compound from the dimethoxyphenethyl series (CID3377343) (1.4 μM). Several other combinations tested with various amides and benzyl substituents showed a range of activity.
Overall, compounds containing an aromatic amide and a benzylic amine substituent substituted at the 3- or 4-position resulted in the highest activities. The probe compound provided the best combination of potency without toxicity and that is why it was selected. All the compounds in this series showed excellent solubility (greater than 100 μM; therefore, this probe (CID3238551/ML158) will serve as a useful probe for cell biology research.
See Appendix C for 1H NMR spectra and LC-MS chromatograms of the probe (CID3238551/ML158) and analogs (CID5113379, CID46897906, CID3377343, CID46912157, CID3500831, CID16470923, CID16470922, CID46912177, CID3237704, CID2930928, CID46897916, CID4207918, CID3422467, CID2838441).
Figure 7 shows the activity of the series of analogs in T. cruzi inhibition assays.
3.5. Cellular Activity
As the primary assay is a phenotypic, cell-based assay, the low nanomolar (nM) activity of the probe (CID3238551/ML158) as described above by definition indicates excellent permeability and good physical properties.
3.6. Profiling Assays
Not applicable.
4. Discussion
4.1. Comparison to Existing Art and Feature/Benefits of the New Probe
The current state of the art for probe molecules in cell culture assays against T. cruzi is a potency of about 1 μM (6), whereas our new probe (CID3238551/ML158) is much more potent at 109 nM, a 10-fold improvement. Also, the current treatments for Chagas disease are toxic to host cells. The probe (CID3238551/ML158) is inactive against host cells at the highest tested dose (19.5 μM), a difference of greater than 500-fold compared with its activity against T. cruzi. These features should make this probe (CID3238551/ML158) very useful as a probe molecule in cell assays and potentially facilitate future in vivo studies.
Investigation into relevant prior art entailed searching the following databases: SciFinder, Patent Lens, PubChem, and PubMed. The search terms applied and hit statistics are provided in Table 4. Abstracts were obtained for all references returned and were analyzed for relevance to the current project. The searches were performed on and are current as of February 16, 2011.
Table 4
Search Strings and Databases Employed in the Prior Art Search.
4.2. Mechanism of Action Studies
In order to elucidate how this probe (CID3238551/ML158) works, an immunofluorescent assay was employed to determine its mode of action and the extent of inhibition of T. cruzi replication in the host cell. To quantitate relative inhibition of parasite replication, 50 infected cells were selected for analysis for the probe (CID3238551/ML158) and each analog. The number of infected cells with more than four amastigotes (where the parasite has invaded and is actively replicating) was divided by the total number of infected cells. As observed by immunofluorescence, all compounds in this series inhibited T. cruzi replication compared with control, but the parasite maintained defined amastigote bodies. Therefore, the mode of action was determined as static. No diffuse staining in the cytosol of host NIH/3T3 cells was observed in any case, suggesting no lysis of the amastigotes as described in Bettiol et al. (7). Therefore, none of the compounds were categorized as lytic.
Additional experiments could be developed to identify the actual molecular target of this probe. A phenotypic cell-based screening approach is extremely powerful for identifying novel compounds of interest when the exact mechanisms of parasite growth are not fully understood. At the Broad Institute, we have developed a robust, scalable method for confident identification of the protein targets of small molecules in their cellular context called stable isotope labeling by amino acids in cell culture (SILAC). SILAC-based target identification technology overcomes prior difficulties with affinity-based target identification methods. This technology is routinely applied at the Broad Institute to identify targets of a variety of small molecules with drug-like properties, including kinase inhibitors, immunophilin modulators, and others.
Another method for target identification would be to develop a T. cruzi strain resistant to the probe (CID3238551/ML158). In brief, T. cruzi could be co-cultured with the probe and compound-resistant parasites would be identified after culturing through many passages. A genomic approach would then be undertaken to identify the gene(s) that differed in the resistant strain versus the wild type. The identification of common mutated genes across multiple experiments could narrow the list of possible targets. Biochemical and biophysical approaches could then be applied for further characterize and optimize the probe (CID3238551/ML158) for in vivo testing.
4.3. Planned Future Studies
The probe (CID3238551/ML158) will be tested for its inhibition of related parasites, Leishmani major and Leishmania amazonensis, with the aim of developing a treatment for Leishmania infections.
The probe (CID3238551/ML158) and certain analogs are also being tested in a mouse model of Chagas disease using fluorescently tagged T. cruzi. Compounds are being dosed intravenously and the extent of T. cruzi replication is assessed by relative fluorescence in the infected area. These studies are ongoing at the University of Georgia by Drs. A. Rodriguez and R. Tarleton.
5. References
- 1.
- De Souza W. Basic cell biology of Trypanosoma cruzi. Curr Pharm Des. 2002;8:269–285. PubMed. [PubMed: 11860366]
- 2.
- Tarleton RL, Reithinger R, Urbina JA, Kitron U, Gürtler RE. The challenges of Chagas Disease: grim outlook or glimmer of hope. PLoS Med. 2007 Dec 27;4(12):e332. [PMC free article: PMC2222930] [PubMed: 18162039] [CrossRef]
- 3.
- WHO Special Program for Research and Training in Tropical Diseases. Report of the Scientific Working Group on Chagas Disease. World Health Organization; 2005.
- 4.
- Konkle ME, Hargrove TY, Kleshchenko YY, von Kries JP, Ridenour W, Uddin MJ, Caprioli RM, Marnett LJ, Nes WD, Villalta F, et al. Indomethacin amides as a novel molecular scaffold for targeting Trypanosoma cruzi sterol 14α-demethylase. J Med Chem. 2009;52(9):2846–2853. PubMed. [PMC free article: PMC2744100] [PubMed: 19354253]
- 5.
- Mott BT, Ferreira RS, Simeonov A, Jadhav A, Ang KK, Leister W, Shen M, Silveira JT, Doyle PS, Arkin MR, et al. Identification and optimization of inhibitors of trypanosomal cysteine proteases: cruzain, rhodesain, and TbCatB. J Med Chem. 2010;53:52–60. PubMed. [PMC free article: PMC2804034] [PubMed: 19908842]
- 6.
- Brak K, Kerr ID, Barrett KT, Fuchi N, Debrath M, Ang K, et al. Nonpeptidic tetrafluorophenoxymehtyl ketone cruzain inhibitors as promising new leads for Chagas disease chemotherapy. J Med Chem. 2010;53:1763–1773. PubMed. [PMC free article: PMC2838180] [PubMed: 20088534]
- 7.
- Bettiol E, Samanovic M, Murkin AS, Raper J, Buckner F, Rodriguez A. Identification of three classes of heteroaromatic compounds with activity against intracellular Trypanosoma cruzi by chemical library screening. PLoS Negl Trop Dis. 2009;3(2):e384. Epub 2009 Feb 24. PubMed. [PMC free article: PMC2639639] [PubMed: 19238193] [CrossRef]
- 8.
- Buckner FS, Verlinde CL, La Flamme AC, Van Voorhis WC. Efficient technique for screening drugs for activity against Trypanosoma cruzi using parasites expressing beta-galactosidase. Antimicrob Agents Chemother. 1996;40(11):2592–2597. PubMed. [PMC free article: PMC163582] [PubMed: 8913471]
Appendix A. Assay Summary Table
Table A1Summary of Completed Assays and AIDs
| PubChem AID No. | Type | Target | Concentration Range (μM) | Samples Tested |
|---|---|---|---|---|
| 1885 | Primary | T. cruzi co-culture | Average 3.75 μM | 303,224 |
| 2044 | Primary | T. cruzi co-culture | 100 nM-6.0 μM | 4063 |
| 2294 | Primary | T. cruzi co-culture | 4.0 nM-1.0 μM | 319 |
| 2630 | Primary | T. cruzi co-culture | 0.11 nM-19.5 μM | 42 |
| 2396 | Primary | T. cruzi co-culture | 0.11 nM-6.8 μM | 27 |
| 493197 | Primary | T. cruzi co-culture | 3 nM-19.5 μM | 46 |
| 2010 | Secondary | NIH3T3 Toxicity | 100 nM-6.0 μM | 4,063 |
| 2586 | Secondary | NIH3T3 Toxicity | 0.11 nM-19.5 μM | 27 |
| 493247 | Secondary | NIH3T3 Toxicity | 3 nM-19.5 μM | 46 |
| 2632 | Orthogonal | Inhibition of T. cruzi replication | Single dose; 10x IC50 as defined in AID 2204 and 2294 | 27 |
| 1968 | Summary | NA | NA | NA |
NA= Not applicable
Appendix B. Detailed Assay Protocols
T cruzi Inhibition Assay Protocol(AID Nos.1885, 2044, 2294, 2630)
Cell Propagation
- 90% DMEM+Phenol red, 10% FBS, 1% PSG.
- Mix and filter through 0.2 microns membrane.
- Keep at 4°C. Warm up to 37°C in water bath before use.
T cruzi Culture and Assays
- 98% DMEM+Phenol red, 2% FBS, 1% PSG.
- Mix and filter through 0.2 microns membrane.
- Keep at 4°C. Warm up to 37°C in water bath before use.
Cell Culture Protocols
NIH/3T3 Cell Culture
- Aspirate medium.
- Rinse cells with 10 ml PBS/plate.
- Aspirate PBS.
- Add 5 ml of pre-warmed trypsin-EDTA, swirl the dish to make sure the trypsin covers all the cells.
- Place the dishes back in the incubator for 5 min.
- Check that the cells are detaching.
- Add 20 ml of medium, pipette up and down to detach all the cells.
- Dilute into 2% FBS/DMEM for assay or 10% FBS/DMEM for propagation.
LLC-MK2 Cell Culture
- Aspirate medium.
- Rinse cells with 10 ml PBS/plate.
- Aspirate PBS.
- Add 5 ml of pre-warmed trypsin-EDTA, swirl the dish to make sure the trypsin covers all the cells.
- Place the dishes back in the incubator for 5 min.
- Check that the cells are detaching.
- Add 20 ml of medium, pipette up and down to detach all the cells.
- Dilute into 2% FBS/DMEM for assay or 10% FBS/DMEM for propagation.
T. cruzi β-gal (Tc) Parasite Culture
- The day before infection (or at least 2–3 hours before), plate 3 million LLC-MK2 cells/T175 in DMEM+10% FBS.
- Either thaw T. cruzi in from liquid nitrogen stock into 50 ml 2% FBS/DMEM or remove the media from a propagating LLC-MK2 co-culture; in other words, harvesting the medium containing the free T. cruzi in 50-ml Falcon tubes.
- Spin 10 min at 2200 rpm.
- Aspirate the supernatant until 15 ml is left.
- Place back the tubes in the incubator delicately (so as not to disturb the pellet).
- Incubate minimum 3 hours at 37°C.
- Take supernatant to fresh tube.
- To count: Mix 75 ml T. cruzi with 25 ml 16% PFA, mix, put 75 ml on Nexcelom cellometer cassette.
- Wait for 2–5 min to let the T. cruzi settle; use the T. cruzi method saved on the M10 machine, press display image, focus, and count.
- Aspirate LLC-MK2 media (used 10% for plating) and replace with 2% FBS/DMEM.
- Plate 17–35 million on T. cruzi on fresh LLC-MK2.
- Change medium after 2 days (Use 2% FBS/DMEM).
- Harvest T. cruzi trypomastigotes on Day 6 and Day 7.
Growth Inhibition Assay Protocol for HTS (384-well plates)
- Warm up medium 2% FBS/DMEM.
- Harvest parasites in 50-ml tubes (1 flask per tube).
- Spin 10 min at 2200 rpm.
- Aspirate media approximately 15 ml and place them on a rack in the incubator to let trypomastigotes swim out of the pellet for 3–5 hours.
- In the meantime, trypsinize NIH/3T3 cells as described in cell culture protocol.
- When NIH/3T3 are detached, harvest them in DMEM- 2% FBS and 1% PSG and count using the NIH3T3 program using the Nexcelom Cellometer.
- Dilute cells to 166,667 cells/ml and add to flask.
- Plate 5,000 cells/30 μL per well using standard cassette multiwell drop Combi, adding at a fast speed.
- Put back in incubator for 3 hours to allow cells to attach.
- Count T. cruzi as described in the parasite culture protocol.
- Dilute to 0.250 million/ml and transfer to a 2-liter flask.
- Add to a 2- liter flask, stirring.
- Pin 50 nL compounds/DMSO to each well with NIH/3T3s.
- Add shortly after 20 μL per well of parasites (5000 T. cruzi) with standard cassette multiwell drop Combi on slow speed.
- Incubate for 4 days (or minimum 90 hours).
- Prepare Gal-Screen with 0.05% NP40.
- Add 30 μL per well of 384-well plate.
- Incubate for 60 minutes.
- Read using the ultrasensitive luminescence program on the Envision (Perkin-Elmer, 0.1 sec/well).
Cell Toxicity Assay Protocol: NIH/3T3 Cells (AID Nos. 2010, 2586, 493247)
- Warm up medium 2% FBS/DMEM.
- Trypsinize NIH/3T3 cells as described in cell culture protocol.
- When NIH3T3 cells are detached, harvest them in DMEM- 2% FBS and 1% PSG and count using the NIH3T3 program using Cellometer Auto T4 (Nexcelom Biosciences).
- Dilute cells to 166,667 cells/ml and add to flask.
- Plate 5,000 cells/50 μL per well using a Thermo MultiDrop Combi liquid dispenser and a sterilized standard sized dispensing cassette adding at a fast speed in a tissue culture hood.
- Put back in incubator for 3 hours to allow cells to attach.
- Pin 50 nL compounds/DMSO to each well.
- Incubate for 4 days (or minimum 90 hours).
- On day of substrate addition, prepare CellTiter-Glo.
CellTiter-Glo® Protocol
- Thaw the CellTiter-Glo Buffer, and equilibrate to room temperature prior to use. For convenience the CellTiter-Glo Buffer may be thawed and stored at room temperature for up to 48 hours prior to use.
- Transfer the appropriate volume of CellTiter-Glo Buffer into the amber bottle containing CellTiter-Glo Substrate to reconstitute the lyophilized enzyme/substrate mixture.
- Mix by gently vortexing, swirling, or by inverting the contents to obtain a homogeneous solution.
- Dilute solution 1:3 with PBS.
- Take out plates from incubator and incubate at r.t. for 30 minutes.
- Add 30 μL/well using a Thermo MultiDrop Combi liquid dispenser and a sterilized standard sized dispensing cassette adding at a fast speed.
- Allow the plate to incubate at room temperature for 10 minutes.
- Read using the ultrasensistive luminescence program on the Envision (Perkin Elmer, 0.1 sec/well).
Intracellular T. cruzi Immunofluorescence Assay Protocol (AID No. 2632)
- Seed NIH/3T3 cells on sterile glass coverslips in 12-well plates and allow cells to adhere overnight.
- Add T. cruzi parasites (MOI 100:1) and allow to infect for 2 h in DMEM+2% FBS and PSG.
- Incubate infected cells for 4 days and fix for 15 min with 4% paraformaldehyde.
- Rinse fixed cells on coverslips with PBS and permeabilize for 15 min in PBS with 0.1% Triton X-100.
- Block for 20 min in PBS with 10% goat serum, 1% bovine serum albumin (BSA), 100 mM glycine, and 0.05% sodium azide.
- Incubate cells for 1 h at room temperature with a polyclonal rabbit anti-T cruzi at 1:2000 dilution.
- Rinse, then add an Alexa Fluor 488 goat anti-rabbit IgG secondary antibody (Molecular Probes, Invitrogen) for 1 h at a 1:800 dilution.
- Stain DNA with DAPI, and mount coverslips with anti-fade mounting media.
- Take images using an inverted Olympus IX70 microscope with a 60X oil objective.
Appendix C. NMR and LC Data of Probe and Analogs

1H NMR (300 MHz, DMSO-d6) Spectrum of the Probe (CID3238551/ML158)

LC-MS Chromatogram of the Probe (CID3238551/ML158)

1H NMR (CDCl3) Spectrum of Analog CID3377343

LC-MS Chromatogram for Analog CID3377343
1H NMR (CDCl3) Spectrum of Analog CID46897906

LC-MS Chromatogram of Analog CID 46897906

1H NMR (300 MHz, DMSO-d6) Spectrum for Analog CID 46912157

LC-MS Chromatogram for Analog CID 46912157

1H NMR (300 MHz, DMSO-d6) Spectrum for Analog CID3500831
LC-MS Chromatogram for Analog CID3500831
1H NMR (300 MHz, DMSO-d6) Spectrum for Analog CID16470923

LC-MS Chromatogram for Analog CID16470923

1H NMR (CDCl3) Spectrum for CID46912177

LC-MS Chromatograms for Analog CID46912177

1H NMR (300 MHz, DMSO-d6) Spectrum for Analog CID 3237704
LC-MS Chromatograms for Analog CID3237704
1H NMR Spectrum (300 MHz, DMSO-d6) for Analog CID87556794
LC-MS Chromatogram for Analog CID87556794

1H NMR (300 MHz, DMSO-d6) Spectrum for Analog CID46897916

LC-MS Chromatogram for Analog CID46897916
1H NMR (300 MHz, DMSO-d6) Spectrum for Analog CID4207918
LC-MS Chromatogram for Analog CID4207918
1H NMR (CDCl3) Spectrum for Analog CID99351072

LC-MS Chromatogram for Analog CID99351072

1H NMR (300 MHz, DMSO-d6) Spectrum for Analog CID87556795

LC-MS Chromatogram for Analog CID87556795
Appendix D. Compounds Submitted to BioFocus
Table A2Probe and Analog Information
| BRD | SID | CID | P/A | MLSID | ML |
|---|---|---|---|---|---|
| BRD-A98248982-001-04-9 | 87219048 | 3238551 | P | MLS002725691 | ML158 |
| BRD-A99518825-001-01-1 | 87556801 | 4207918 | A | MLS002725685 | NA |
| BRD-A83402799-001-01-8 | 87219049 | 3500831 | A | MLS002725682 | NA |
| BRD-A74871599-001-01-7 | 87556793 | 3237704 | A | MLS002725692 | NA |
| BRD-A37656721-001-01-9 | 87556802 | 44629413 | A | MLS002725684 | NA |
NA- Not applicable; P = probe; A = analog
- PMCPubMed Central citations
- PubChem SubstanceRelated PubChem Substances
- PubMedLinks to PubMed
- Review Identification of Small-Molecule Inhibitors of Trypansoma cruzi Infection - Probe 1.[Probe Reports from the NIH Mol...]Review Identification of Small-Molecule Inhibitors of Trypansoma cruzi Infection - Probe 1.Carmody LC, Germain A, Barker D, Galan-Rodriguez C, Bettiol E, Rodriguez A, MacPherson L, Palmer M, Schreiber SL. Probe Reports from the NIH Molecular Libraries Program. 2010
- Review Identification of Small-Molecule Inhibitors of Trypansoma cruzi Infection.[Probe Reports from the NIH Mol...]Review Identification of Small-Molecule Inhibitors of Trypansoma cruzi Infection.Carmody LC, Germain A, Barker D, Galan-Rodriguez C, Bettiol E, Rodriguez A, MacPherson L, Palmer M, Schreiber SL. Probe Reports from the NIH Molecular Libraries Program. 2010
- Review Identification of Diversity-Oriented Synthesis Derived Small Molecule, ML341, with Cidal Activity Against Trypanosoma cruzi.[Probe Reports from the NIH Mol...]Review Identification of Diversity-Oriented Synthesis Derived Small Molecule, ML341, with Cidal Activity Against Trypanosoma cruzi.Carmody LC, Germain AR, Engel JC, Gut J, Kaiser M, Jewett I, LeQuemen S, Marie JC, Dandapani S, Rodriguez A, et al. Probe Reports from the NIH Molecular Libraries Program. 2010
- Identification of small-molecule inhibitors of Trypansoma cruzi replication.[Bioorg Med Chem Lett. 2011]Identification of small-molecule inhibitors of Trypansoma cruzi replication.Germain AR, Carmody LC, Dockendorff C, Galan-Rodriguez C, Rodriguez A, Johnston S, Bittker JA, MacPherson L, Dandapani S, Palmer M, et al. Bioorg Med Chem Lett. 2011 Dec 1; 21(23):7197-200. Epub 2011 Sep 29.
- Compound profiling and 3D-QSAR studies of hydrazone derivatives with activity against intracellular Trypanosoma cruzi.[Bioorg Med Chem. 2016]Compound profiling and 3D-QSAR studies of hydrazone derivatives with activity against intracellular Trypanosoma cruzi.Costa LB, Cardoso MV, de Oliveira Filho GB, de Moraes Gomes PA, Espíndola JW, de Jesus Silva TG, Torres PH, Silva Junior FP, Martin J, de Figueiredo RC, et al. Bioorg Med Chem. 2016 Apr 15; 24(8):1608-18. Epub 2016 Feb 23.
- Identification of Small-Molecule Inhibitors of Trypansoma cruzi Infection - Prob...Identification of Small-Molecule Inhibitors of Trypansoma cruzi Infection - Probe 3 - Probe Reports from the NIH Molecular Libraries Program
Your browsing activity is empty.
Activity recording is turned off.
See more...