NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
This probe (ML172, CID 44602489), which possesses improved muscarinic selectivity over our previous M5 positive allosteric modulator (PAM) probe (ML129, CID 42633508), can be used for in vitro molecular pharmacology and electrophysiology experiments to study the role of selective M5 receptor activation. This probe possesses unprecedented selectivity versus M1, M2, M3 and M4. The probe (ML172, CID 44602489) is not readily CNS penetrant, and so would need to be administered i.c.v. to study the role of central M5 activation in vivo.
Assigned Assay Grant #: MH077607-1
Screening Center Name & PI: Vanderbilt Screening Center for GPCRs, Ion Channels and Transporters, C. David Weaver
Chemistry Center Name & PI: Vanderbilt Specialized Chemistry Center for Accelerated Probe Development, Craig W. Lindsley
Assay Submitter & Institution: P. Jeffrey Conn, Vanderbilt University
PubChem Summary Bioassay Identifier (AID): 2416
Probe Structure & Characteristics
1-(4-phenoxybenzyl)-5-(trifluoromethoxy)indoline-2,3-dione, MW = 413.4, logP = 5.8, tPSA = 107.0 Å2

ML172
| CID/ML# | Target Name | IC50/EC50 (nM) [SID, AID] | Anti-target Name(s) | IC50/EC50 (μM) [SID, AID] | Fold Selective | Secondary Assay(s) Name: IC50/EC50 (nM) [SID, AID]§ |
|---|---|---|---|---|---|---|
| CID 44602489/ML172 | M5 | 1900 [SID 87352032, AID 2416] | M1, M2, M3, M4 | > 30 μM [SID 87352032, AID 2204] | > 30 | ACh Fold-Shift (5-fold) [SID 87352032, AID 2186] |
Recommendations for Scientific Use of the Probe
This probe (ML172, CID 44602489), which possesses improved muscarinic selectivity over our previous M5 PAM probe (ML 129, CID 42633508), can be used for in vitro molecular pharmacology and electrophysiology experiments to study the role of selective M5 receptor activation. This probe possesses unprecedented selectivity versus M1, M2, M3 and M4. The probe (ML172, CID 44602489) is not readily CNS penetrant, and so would need to be administered i.c.v. to study the role of central M5 activation in vivo.
1. Introduction
Specific AIM: To identify small molecule positive allosteric modulators (PAMs) and/or allosteric agonists of the M5 muscarinic acetylcholine receptor that are cell permeable, possess low- to sub-micromolar potency and show greater than 10-fold selectivity over the other mAChRs (M1, M2, M3 and M4) employing a functional HTS approach. Out of this effort primarily aimed at M1, which afforded a highly selective M1 allosteric agonist (CID 25010775) and two highly selective M1 PAM probes (CID 44251556, and CID 44475955), we also identified and optimized the first selective M5 PAM (CID 42633508), and now report the next-generation M5 PAM described herein. Another MLSCN screening effort identified a highly selective M4 PAM (CID 864492) and a subsequent highly selective M4 PAM endowed with improved human potency (ML 173, CID 45142486); thus, a toolkit containing highly selective, allosteric mAChR ligands are available from the MLPCN to study individual mAChR function.
Significance: The five cloned muscarinic acetylcholine receptor subtypes (mAChR1-5 or M1–5) are known to play highly important and diverse roles in many basic physiological processes.1–3 Correspondingly, muscarinic agonists and antagonists targeting one or more subtypes have been used preclinically and clinically for research and treatment of a wide range of pathologies.3,4 Based on the high sequence homology of the mAChRs across subtypes, and particularly within the orthosteric acetylcholine (ACh) binding site, discovery of truly subtype-selective compounds has proven historically difficult. Due to the scarcity of selective compounds, a detailed understanding of the precise roles of each subtype in neurobiology and in various central nervous system (CNS) disorders has thus remained challenging.3,4 A number of novel highly subtype-selective allosteric ligands for M1 and M4 have emerged from functional cell-based screening efforts.5,6 Only recently have the first M5 selective ligands been disclosed from these laboratories.13 Relative to the other mAChRs, little is known about M5, which is expressed at very low levels in the CNS and peripheral tissues.2–4
Rationale: Data from studies using mAChR5 knock-out (M5-KO) mice suggest that M5 is the sole mediator of ACh-induced vasodilation in the cerebral vasculature and thereby may have therapeutic relevance for cerebrovascular diseases or acute ischemic stroke.7,8 M5-KO mice have also been found to exhibit deficits in long-term potentiation (LTP) at the hippocampal mossy fiber-CA3 synapse and show deficits in hippocampal-dependent behavioral cognitive tests.8 In light of these and related findings, activation of M5 has been suggested as a potential target for treatment of Alzheimer’s disease, perhaps in combination with M1 activation.9 Consistent with the putative post-synaptic localization of M5 in the ventral tegmental area (VTA), other M5-KO data suggest this subtype plays an important role in regulation of mesolimbic dopamine transmission.3,9 Indeed, M5-KO mice exhibit decreased reward responses to morphine, decreased self-administration of cocaine, and less pronounced drug withdrawal symptoms, suggesting that M5 antagonists or negative allosteric modulators may have therapeutic value in the treatment of illicit drug addiction.9–11 Further pharmacological exploration of these and related hypotheses greatly depends on the discovery of novel M5-preferring or selective small molecule tools.
2. Materials and Methods
2.1. Assays
PubChem Primary Assay Description: Chinese hamster ovary (CHO K1) cells stably expressing rat (r)M1 were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and cultured according to their recommendations. CHO cells stably expressing human (h) M2, hM3, and hM5 were generously provided by A. Levey (Emory University, Atlanta, GA); rM4 cDNA provided by T. I. Bonner (National Institutes of Health, Bethesda, MD) was used to stably transfect CHO-K1 cells purchased from the ATCC using Lipofectamine 2000. To make stable hM2 and rM4 cell lines for use in calcium mobilization assays, cell lines were cotransfected with a chimeric G protein (Gqi5) using Lipofectamine 2000. rM2, hM3, and hM5 cells were grown in Ham’s F-12 medium containing 10% heat-inactivated fetal bovine serum, 2 mM GlutaMax I, 20 mM HEPES, and 50 μg/mL G418 sulfate. hM2-Gqi5 cells were grown in the same medium supplemented with 500 μg/mL hygromycin B. Stable rM4 cells were grown in Dulbecco’s modified Eagle’s medium containing 10% heat-inactivated fetal bovine serum, 2 mM GlutaMax I, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 20 mM HEPES, and 400 μg/mL G418 sulfate; rM4-Gqi5 cells were grown in the same medium supplemented with 500 μg/mL hygromycin B. CHO cells stably expressing rM1, hM3, or hM5 were plated at a seeding density of 50,000 cells/100 μL/well. CHO cells stably coexpressing hM2/Gqi5 and rM4/Gqi5 were plated at a seeding density of 60,000 cells/100 μL/well. For calcium mobilization, cells were incubated in antibiotic-free medium overnight at 37 °C/5% CO2 and assayed the next day.
Calcium Mobilization Assay: Cells were loaded with calcium indicator dye [2 μM Fluo-4 acetoxymethyl ester (50 μL/well) prepared as a stock in DMSO and mixed in a 1:1 ratio with 10% Pluronic acid F-127 in assay buffer (1xHanks’ balanced salt solution supplemented with 20 mM HEPES and 2.5 mM probenecid, pH 7.4)] for 45 min at 37 °C. Dye was removed and replaced with the appropriate volume of assay buffer. All compounds were serially diluted in assay buffer for a final 2x stock in 0.6% DMSO. This stock was then added to the assay plate for a final DMSO concentration of 0.3%. Acetylcholine (EC20 concentration or full dose-response curve) was prepared at a 10x stock solution in assay buffer before addition to assay plates. Calcium mobilization was measured at 25 °C using a FLEXstation II (Molecular Devices, Sunnyvale, CA). Cells were preincubated with test compound (or vehicle) for 1.5 min before the addition of the agonist, acetylcholine (ACh). Cells were then stimulated for 50 s with a submaximal concentration (EC20) or a full dose-response curve of acetylcholine. The signal amplitude was first normalized to baseline and then as a percentage of the maximal response to acetylcholine.
2.2. Probe Chemical Characterization
Synthetic procedure (large scale) and spectral data for ML172 (CID 44602489)
Probe compound ML172 (CID 44602489) was prepared according to the above scheme and provided the following characterization data: LCMS (>98%) m/z = 414 [M+H+] (1.65 min retention, 214 nm),1H NMR (>95%) (400 MHz, CDCl3) δ =7.52 (s, 1H), 7.36 (m, 6H), 7.15 (t, J = 7.2 Hz, 1H), 7.10 (t, J = 5.2 Hz, 2H), 6.86 (d, J = 8.8 Hz, 2H), 4.93 (s, 2H). 13C-NMR (100MHz, d6 - DMSO) δ 182.2, 157.8, 157.6, 156.4, 145.3, 130.9, 129.8, 129.0, 128.3, 123.7, 121.6, 120.4, 119.2, 119.0, 118.5, 118.1, 111.9, 43.7, HRMS (Q-TOF): m/z calc for C22H15NO4F3 [M + H]: 414.0953, found 414.0959.
Solubility. Solubility in PBS (at pH = 7.4) was determined to be less than 0.10 μM although alternative formulations could potentially improve this number. For example, ML172 shows good solubility in DMSO (>100 μM), but this is not a universally acceptable vehicle.
Stability. Stability (at room temperature = 23 °C) for ML172 in PBS (no antioxidants or other protectorants and DMSO concentration below 0.1%) is shown in the table below. After 48 hours, the percent of parent compound remaining was 134%, but the assay variability over the course of the experiment ranged from a low of 84% (at 24 hours) to a high of 167% (at 30 minutes). It is important to point out that ML172 showed very poor LCMS sensitivity and exhibited high levels of non-specific binding. Given these caveats, it is difficult to draw any firm conclusions with respect to ML172’s stability from this experiment.
| Percent Remaining (%) | |||||||
|---|---|---|---|---|---|---|---|
| Compound | 0 Min | 15 Min | 30 Min | 1 Hour | 2 Hour | 24 Hour | 48 Hour |
| ML172 | 100 | 164 | 167 | 154 | 114 | 84 | 134 |
Reactivity. As assessed through a glutathione (GSH) trapping experiment in phosphate buffered saline (with a substrate concentration of typically 5–50 μM and a GSH concentration of 5 mM, at t = 60 minutes) ML172 was found to not form any detectable GSH adducts.21
Compounds added to the SMR collection (MLS#s): 003108019 (ML172, CID 44602489, 500 mg), 003108020, 003108021, 003108022, 003108023, 003108024.
2.3. Probe Preparation
1-(4-phenoxybenzyl)-5-(trifluoromethoxy)indoline-2,3-dione (ML172, CID 44602489): To a flask containing acetonitrile (20 mL) was added 5-trifluoromethoxyisatin (750 mg, 3.245 mmol), potassium carbonate (897 mg, 6.490 mmol), 4-phenoxybenzyl bromide (897 mg, 3.407 mmol), and potassium iodide (54 mg, 0.325 mmol). The reaction was stirred overnight at room temperature and then judged complete by LCMS analysis. The solution was partitioned between CH2Cl2 and H2O, the organics were dried over MgSO4, and then the solution was filtered and concentrated in vacuo to afford the crude product. Purification by silica gel plug (1:3 EtOAc:Hexanes fixed solvent gradient) afforded the title compound as a bright red-orange sticky solid (1.340 g, 3.242 mmol, 99%)
3. Results
Center Summary of Screen: This screen was performed in the pilot phase, the MLSCN, when the MLSMR compound collection at Vanderbilt only contained 65K compounds. From the primary M1 screen of 65K compounds, ~12 putative M1 PAMs were identified giving an average Z’ score of 0.70±0.09. The confirmation screen (singles at 10 μM) produced two lead compounds, one of which was recently developed into a structurally novel M1 PAM (CID 44475955). The other, CID 3008304, represented a unique, and never before seen, pharmacological profile (Figure 1) in that it was a PAM of all the Gq-coupled mAChRs (M1 EC50 = 6.1 μM, M3 EC50 = 6.4 μM and M5 EC50 = 4.1 μM), but devoid of activity at the Gi/o-coupled M2 and M4.12 This led us to predict it would be possible to dial-in or dial-out different mAChR sub-types and potentially develop an M1 selective PAM, an M3 selective PAM and/or an M5 selective PAM from this non-selective lead through chemical optimization.13a As predicted, CID 3008304 served as the common starting point for the development of the two highly selective probe molecules shown in Figure 2: the M1 PAM (CID 44251556) and the M5 PAM (CID 42633508, EC50 = 1.1 μM, 91% ACh Max). Although very selective for M5, CID 42633508 did retain a detectable amount of activity at the M1 and M3 receptors at concentrations of 30 μM and above. Therefore we chose to explore the possibility of further optimizing CID 42633508 as described next.

Figure 1
CRCs at M1–M5 for HTS lead CID 3008304. CID 3008304 is a pan Gq-M1, M3, M5 PAM

Figure 2
Two MLPCN probe molecules derived from CID 3008304.
Probe Optimization Strategy: For the continued optimization of CID 42633508, we chose the strategy depicted in Figure 3, and as SAR with allosteric ligands is often shallow, we employed an iterative parallel synthesis approach. From the development of our initial M5 probe, we were confident that the 5-OCF3 group was essential for M5-preferring activity and selectivity, so this moiety was universally retained. Libraries were prepared according to Scheme 1, wherein the commercially available indoline-2,3-dione 1 was alkylated with 4-bromobenzylbromide to deliver intermediate 2. An eleven-membered Suzuki library was then prepared to explore the biaryl and heterobiaryl replacements for the methyl ether of CID 42633508, thus providing analogs 3. Concomitantly, 1 was alkylated with functionalized phenethyl bromides 4 to explore the effect of chain homologation present in analogs 5. Compounds from these libraries were then triaged by a single point (10 μM) screen for their ability to potentiate an EC20 of ACh in M5-CHO cells with those results appearing in Figure 4.

Figure 3
Optimization strategy for CID 42633508, a highly M5-preferring PAM.

Scheme 1
Reagents and conditions. (a) p-bromobenzylbromide, K2CO3, KI, CAN, rt, 16 h (99%); (b) R-B(OH)2, Pd(t-Bu3P)2, Cs2CO3, THF:H2O, mw, 120 °C, 20 min (10–90%); (c) K2CO3, KI, CAN, rt, 16 h (50–90%)
In general, chain homologation in analogs 5 did not produce improved profiles relative to the benzyl analogs 3. Although M5 PAM efficacy was maintained it came at the cost of right-shifted EC50 values. For example, CID 45281805, the direct phenethyl analog of CID 42633508 possessed an M5 EC50 of 4.9 and an 80% ACh maximum response (data not shown). Biaryl and heterobiaryl analogs 3 proved far more productive, affording a number of M5 PAMs with high selectivity versus M1 (M1 EC50s >30 μM) and low micromolar M5 EC50s (Table 1). The remaining 5 analogs of 3 displayed M5 EC50s > 10 μM. A wide variety of R- groups could be tolerated, both five- (CIDs 45281797 and 45281803) and six-membered heterocycles (CIDs 45281798 and 45281801) provided active compounds, as did bare phenyl (CID 45281794) and substituted phenyl (CID 45281796). Potency was virtually identical for all of the active analogs 3 (M5 EC50s 2.7 – 4.8 μM) with similar ACh Max values (70–85%). Shallow SAR was again noted for these allosteric ligands, with compounds either being active in the micromolar potency range or inactive as M5 PAMs.
Table 1
Structures and activities of analogs 3.
The two most potent analogs from Table 1 (CIDs 45281794 and 45281797) were selected for additional follow-up. Figure 5 depicts only the Gq mAChR (M1, M3 and M5) concentration response curves (CRCs) for CID 45281794 and CID 45281797. Note that CID 45281794 possesses improved M5 selectivity versus our initial M5 probe CID 42633508, with almost negligible activation of M3 at 30 μM (Figure 5A). Both analogs CID 45281794 and CID 45281797 elicit significant leftward shifts (> 50-fold) of the ACh CRC at M5 (Figure 5C), as compared to the 14-fold shift of our initial M5 probe CID 42633508. As seen with the M1 PAM BQCA14–16 and other ago-potentiators for class C GPCRs.17–19 Figure 5C indicates moderate intrinsic allosteric agonism at 30 μM for both analogs at the M5 receptor.
Encouraged by the potency and mAChR selectivity of the phenyl-containing CID 45281794 and the ether-containing CID 42633508 (Figure 2), a hydrid analog was prepared (Scheme 2) that possessed a biphenyl ether moiety, CID 44602489 (ML 172), and displayed an M5 EC50 of 1.9 μM with a 75% ACh Max. More Importantly, CID 44602489 (ML 172) was completely selective versus M1–4, affording no elevation of an ACh EC20 at M1–4 at concentrations as high as 30 μM (Figure 6). Notably, CID 44602489 (ML 172) represents the most selective M5 PAM described to date; however, unlike CID 45281794 and CID 45281797, this hybrid analog only afforded a ~5-fold shift of the M5 ACh CRC at 30 μM and did not display intrinsic allosteric agonism (graph not shown). While subsequent compounds were prepared in an effort to improve this fold shift, potency and fold shift do not always track in the same direction, and in this case our attempts were unfruitful. Therefore CID 44602489 (ML172), the most highly M5 muscarinic selective PAM known to date, was chosen as our next-generation M5 probe molecule.

Scheme 2
Reagents and conditions: (a) K2CO3, KI, CAN, mw, 160 °C, 10 min (68%).
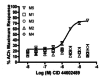
Figure 6
Concentration-response curves in Ca2+ mobilization assays with M1-,M2Gqi5-, M3, M4/Gqi5-, and M5-CHO cells for CID 44602489 (M5 EC50 = 1.9 μM). Data represent means ± SEM from 3 independent determinations
Subsequent follow up demonstrated that many of these analogs displayed moderate to poor PK in rats with limited brain exposure (AUCBrain/AUCPlasma ~ 0.25), presumably due to the bis-carbonyl of the isatin moiety. However, these are important tools to study M5 function in cells, in electrophysiology and by i.c.v. injection. We did not examine the brain exposure when a DMSO-containing vehicle was employed, but using a DMSO-containing vehicle has been shown to improve brain levels.17–18
The calculated physical properties appearing in Table 2 for the in initial M5 probe molecule (CID 42633508) and this current probe ML172 (CID 44602489) were generated using TRIPOS software. Also included in Table 2 are the averages from the MDDR database of compounds both entering Phase I and launched drugs. These numbers indicate that both probes are within the average values for Phase I compounds and launched compounds with respect to MW, hydrogen bond donors/acceptors and number of rotatable bonds. However, both molecules have relatively high cLogP values, which may contribute to their moderate to poor PK, and is also supported by the calculated low LogS (Solubility) values of −4.94 and −6.67 (CID 42633508 and CID 44602489, respectively). Similarly, the greater than 100 square angstroms of polar surface area for each molecule, although not necessarily high enough to preclude entry into the CNS, might be contributing to their less than ideal CNS exposure.
Table 2
Calculated Property Comparison with MDDR Compounds.
To more fully characterize this novel M5 PAM probe molecule, ML172 (CID 44602489) was tested at MDS Pharma’s (now Ricerca’s) Lead Profiling Screen (binding assay panel of 68 GPCRs, ion channels and transporters screened at 10 μM), and was found to not significantly interact with 36 out of the 68 assays conducted (no inhibition > 50% at 10 μM).20 ML172 (CID 44602489) did have activity at the following 32 targets (human targets at 10 μM, unless stated as rat): Adenosine A1 (50%), Adenosine A3 (97%), rat Adrenergic α1A (79%), Adrenergic α1D (66%), Adrenergic α2A (100%), Adrenergic β1 (89%), NET (93%), Bradykinin B1 (52%), rat Calcium Channel L-Type, Dihydropyridine (74%), CB1 (109%), D1 (99%), D2S (85%), D3 (79%), DAT (90%), rat GABA transporter (56%), H1 (99%), H2 (101%), rat Imidazoline, central (50%), Leukotriene, Cysteinyl CysLT1 (83%), MT1 (60%), Muscarinic M1 (98%), Muscarinic M2 (96%), Neuropeptide Y Y1 (83%), KOP (98%), MOP (99%), PAF (58%), hERG (81%), EP4 (66%), 5-HT1A (69%), 5-HT2B (90%), SERT (80%) and rat Sodium Channel, site 2 (97%). However it should be pointed out that these are only single-point values and that functional selectivity may be significantly better than suggested by these “% activities.” This is best illustrated by the fact that we see no functional activity for ML172 (CID 44602489) at either the M1 or M2 receptor at concentrations as high as 30 μM (Figure 6), while the lead profiling screen indicates 98% and 96% inhibition of radio ligand binding at M1 and M2, respectively, at just 10 μM.
In summary, ML172 (CID 44602489) represents a marked improvement over our initial M5 probe with respect to muscarinic selectivity, however ancillary pharmacology, at the binding level, appears to have suffered. CNS penetration is poor, using DMSO-free vehicles, possibly restricting ML172 (CID 44602489) to use as an in vitro or i.c.v. probe.
3.1. Summary of Screening Results

3.2. Dose Response Curves for Probe
See Figure 6 (vide supra) for M5 PAM potency and selectivity against M1–4.
3.3. Scaffold/Moiety Chemical Liabilities
No chemical liabilities for the probe molecule have been identified at the present time; however its high lipophilicity may be contributing to a less than ideal profile in the Lead Profiling Screen (Ricerca).20 Additionally, the isatin core could be viewed as a liability given historical data. To address this potential issue one could envision various isatin replacements, but given the steep SAR generally associated with allosteric modulators it would be a significant undertaking to embark on such an effort. As such, the replacement of the isatin core might be more adequately undertaken as part of an extended probe characterization proposal.
3.4. SAR Table
Table 3SAR Analysis for M1 Positive Allosteric Modulators
 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Entry | CID | SID | VU Number | * | X | Y | Z | M1 PAM EC50 (μM)** | M1 ACh fold-shift*** |
| 1 | 45281794 | 92845376 | VU0365114-1 | S | methylene | phenyl | - | 2.7 | > 50 |
| 2 | 45281795 | 92845377 | VU0365115-1 | S | methylene | 3,5-dichlorophenyl | - | ~ 10 | ND |
| 3 | 45281796 | 92845378 | VU0365116-1 | S | methylene | phenyl | 2-methoxy | 3.9 | ND |
| 4 | 45281797 | 92845379 | VU0365117-1 | S | methylene | thiophen-3-yl | - | 2.8 | > 50 |
| 5 | 45281798 | 92845380 | VU0365118-1 | S | methylene | pyridin-3-yl | 6-chloro | 4.8 | ND |
| 6 | 45281799 | 92845381 | VU0365119-1 | S | methylene | phenyl | 4-chloro | ~ 10 | ND |
| 7 | 45281800 | 92845382 | VU0365120-1 | S | methylene | 2,5-difluorophenyl | - | ~ 10 | ND |
| 8 | 45281801 | 92845383 | VU0365121-1 | S | methylene | pyridin-3-yl | 6-fluoro | 3.6 | ND |
| 9 | 45281802 | 92845384 | VU0365122-1 | S | methylene | phenyl | 4-methoxy | ~ 10 | ND |
| 10 | 45281803 | 92845385 | VU0365123-1 | S | methylene | 1H-pyrazol-4-yl | 1-methyl | 3.3 | ND |
| 11 | 45281804 | 92845386 | VU0365138-1 | S | methylene | phenyl | 4-propoxy | > 10 | ND |
| 12 | 45281805 | 92845387 | VU0365124-1 | S | ethyl | 4-methoxy | - | 4.9 | ND |
| 13 | 45281806 | 92845388 | VU0365125-1 | S | ethyl | 4-bromo | - | ~10 | ND |
| 14 | 44602489 | 87352032 | VU0400265-1 | S | methylene | O | phenyl | 1.9 | 5 |
3.5. Cellular Activity
This series of positive allosteric modulators displayed functional activity (Ca+2 mobilzation) in CHO cells stably expressing M1–5 receptors [AID 2416 and 2204]. Many of these analogs were tested in rat PK, but were found to give moderate to poor exposure (low absolute levels detected) and limited brain exposure (AUCBrain/AUCPlasma ~ 0.25) demonstrating that at least some level of CNS exposure was obtained.
4. Discussion
4.1. Comparison to existing art and how the new probe is an improvement
Presently there are no known selective M5 PAMs with the exception of those disclosed from our labs, which include the two MLPCN probe molecules (ML 129 and ML172). Relative to our initial M5 PAM probe (ML 129, CID 42633508) this second generation probe (ML172, CID 44602489) possesses dramatically improved selectivity relative to the other M1–4 receptors. The previous probe, although very selective, did begin to show a trend towards off-target receptor activation at the highest doses, ML172 (CID 44602489) is clearly improved in this context (Figure 6). Lastly, this probe (ML172, CID 44602489), along with our initial M5 probe (ML 129, CID 42633508), is not encumbered with patent restrictions and rests firmly in the public domain.
4.2. Mechanism of Action Studies
It is believed that this probe (ML172, CID 44602489) is functioning by positive allosteric modulation of the muscarinic acetylcholine M5 receptor given its lack of activity on the other muscarinic receptor subtypes (M1–4) and its dependence on the presence of ACh to elicit functional activity.
4.3. Planned Future Studies
Future work will focus on developing a more centrally penetrant M5 PAM with improved physical properties, which maintains this unprecedented level of muscarinic selectivity and possesses an improved ancillary pharmacology profile. Ultimately it would be highly desirable to develop such a selective M5 PAM for use as an in vivo tool to better understand the biological processes associated with the M5 receptor.
5. References
- 1.
- Bonner TI, Buckley NJ, Young AC, Brann MR. Science. 1987;237:527–532. [PubMed: 3037705]
- 2.
- Bonner TI, Young AC, Brann MR, Buckley NJ. Neuron. 1988;1:403–410. [PubMed: 3272174]
- 3.
- Wess J. Annu Rev Pharmacol Toxicol. 2004;44:423–450. [PubMed: 14744253]
- 4.
- Langmead CJ, Watson J, Reavill C. Pharmacol Ther. 2008;117:232–243. [PubMed: 18082893]
- 5.
- Birdsall NJM, Lazareno S. Mini Rev Med Chem. 2005;5:523–543. [PubMed: 15974931]
- 6.
- Conn PJ, Christopoulos A, Lindsley CW. Nat Rev Durg Disc. 2009;8:41–54. [PMC free article: PMC2907734] [PubMed: 19116626]
- 7.
- Yamada M, Lamping KG, Duttaroy A, Zhang W, Cui Y, Bymaster FP, McKinzie DL, Felder CC, Deng C, Faraci FM, Wess J. Proc Natl Acad Sci. 2001;98:14096–14101. [PMC free article: PMC61174] [PubMed: 11707605]
- 8.
- Araya R, Noguchi T, Yuhki M, Kitamura N, Higuchi M, Saido TC, Seki K, Itohara S, Kawano M, Tanemura K, Takashima A, Yamada K, Kondoh Y, Kanno I, Wess J, Yamada M. Neurobiol Dis. 2006;24:334–344. [PubMed: 16956767]
- 9.
- Wess J, Eglen RM, Gautam D. Nat Rev Drug Discov. 2007;6:721–733. [PubMed: 17762886]
- 10.
- Basile AS, Fedorova I, Zapata A, Liu X, Shippenberg T, Duttaroy A, Yamada M, Wess J. Proc Natl Acad Sci. 2002;99:11452–11457. [PMC free article: PMC123277] [PubMed: 12154229]
- 11.
- Thomsen M, Woldbye DPD, Wortwein G, Fink-Jensen A, Wess J, Caine SB. J Neurosci. 2005;25:8141–8149. [PMC free article: PMC6725551] [PubMed: 16148222]
- 12.
- Marlo JE, Niswender CM, Days EL, Bridges TM, Xiang Y, Rodriguez AL, Shirey JK, Brady AE, Nalywajko T, Luo Q, Austin CA, Williams MB, Kim K, Williams R, Orton D, Brown HA, Lindsley CW, Weaver CD, Conn PJ. Mol Pharmacol. 2009;75:577–588. [PMC free article: PMC2684909] [PubMed: 19047481]
- 13.
- a. Bridges TM, Marlo JE, Niswender CM, Jones JK, Jadhav SB, Gentry PR, Weaver CD, Conn PJ, Lindsley CW. J Med Chem. 2009;52:3445–3448. [PMC free article: PMC3875304] [PubMed: 19438238]
b. Bridges TM, Kennedy JP, Cho HP, Breinginger ML, Gentry PR, Hopkins CR, Conn PJ, Lindsley CW. Bioorg Med Chem Lett. 2010;20:558–562. [PMC free article: PMC3177601] [PubMed: 20004578] - 14.
- Ma L, Seager M, Wittman M, Bickel N, Burno M, Jones K, Graufelds VK, Xu G, Pearson M, McCampbell A, Gaspar R, Shughrue P, Danzinger A, Regan C, Garson S, Doran S, Kreatsoulas C, Veng L, Lindsley CW, Shipe W, Kuduk S, Jacobson M, Sur C, Kinney G, Seabrook GR, Ray WJ. Proc Natl Acad Sci USA. 2009;106:15950–15955. [PMC free article: PMC2732705] [PubMed: 19717450]
- 15.
- Shirey JK, Brady AE, Jones PJ, Davis AA, Bridges TM, Jadhav SB, Menon U, Christain EP, Doherty JJ, Quirk MC, Snyder DH, Levey AI, Watson ML, Nicolle MM, Lindsley CW, Conn PJ. J Neurosci. 2009;29:14271–14286. [PMC free article: PMC2811323] [PubMed: 19906975]
- 16.
- Yang FV, Shipe WD, Bunda JL, Nolt MB, Wisnoski DD, Zhao Z, Barrow JC, Ray WJ, Ma L, Wittman M, Seager M, Koeplinger K, Hartman GD, Lindsley CW. Bioorg Med Chem Lett. 2010;20:531–536. [PubMed: 20004574]
- 17.
- Lindsley CW, Wisnoski DD, Leister WH, O’Brien JA, Lemiare W, Williams DL Jr, Burno M, Sur C, Kinney GG, Pettibone DJ, Tiller PR, Smith S, Duggan ME, Hartman GD, Conn PJ, Huff JR. J Med Chem. 2004;47:5825–5828. [PubMed: 15537338]
- 18.
- Kinney GG, O’Brien JA, Lemaire W, Burno M, Bickel DJ, Clements MK, Wisnoski DD, Lindsley CW, Tiller PR, Smith S, Jacobson MA, Sur C, Duggan ME, Pettibone DJ, Williams DW Jr. J Pharmacol Exp Ther. 2005;313:199–206. [PubMed: 15608073]
- 19.
- Engers DW, Rodriguez AL, Oluwatola O, Hammond AS, Venable DF, Williams R, Sulikowski GA, Conn PJ, Lindsley CW. ChemMedChem. 2009;4:505–511. [PMC free article: PMC2865690] [PubMed: 19197923]
- 20.
- For information on the Ricerca (formerly MDS Pharma) Lead Profiling Screen see: https:
//pharmacology .ricerca.com/Catalog/ - 21.
- Solubility (PBS at pH = 7.4), Stability and Reactivity experiments were conducted at Absorption Systems. For additional information see: https://www
.absorption.com/site
- PMCPubMed Central citations
- PubChem BioAssay for Chemical ProbePubChem BioAssay records reporting screening data for the development of the chemical probe(s) described in this book chapter
- PubChem SubstanceRelated PubChem Substances
- PubMedLinks to PubMed
- Development of the First Highly Selective mAChR 5 (M5) Positive Allosteric Modul...Development of the First Highly Selective mAChR 5 (M5) Positive Allosteric Modulator (PAM) - Probe Reports from the NIH Molecular Libraries Program
Your browsing activity is empty.
Activity recording is turned off.
See more...

 .
.
