NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
The recently discovered apelin receptor (APJ, AGTRL-1, APLNR) system has emerged as a critical mediator of cardiovascular homeostasis involved in the pathogenesis of hypertension, heart failure, atherosclerosis and other cardiovascular diseases. Herein is presented the discovery and characterization the first non-peptide based potent (3.7 μM) small molecule APJ functional agonist in cell-based assays, that is >21 fold selective over the closely related angiotensin 1 (AT1) receptor, derived from a high throughput screen (HTS) of the ~330,600 compound Molecular Libraries Small Molecule Repository (MLSMR) collection. This agonist showed some binding activity against 4 out of 37 other GPCRs and transporters, including the 5-HT1A, α2C adrenergic, and benzylpiperazine receptors (55%I, 51%I and 65%I at 10 μM, respectively) and the norepinephrine transporter (57%I at 10 μM). The synthetic methodology, development of structure-activity relationships (SAR), and initial in vitro pharmacologic characterization are also presented. This probe molecule provides a useful tool compound for investigators interested in understanding apelin receptor pharmacology and function.
Assigned Assay Grant #: 1R21NS059422-01
Screening Center Name & PI: Sanford Burnham Center for Chemical Genomics (SBCCG) & John C. Reed (PI)
Chemistry Center Name & PI: same as above
Assay Submitter & Institution: Layton H. Smith, Sanford-Burnham Medical Research Institute
PubChem Summary Bioassay Identifier (AID): 2580
Probe Structure & Characteristics
This Center Probe Report describes the first reported small molecule APJ agonist, ML233, which is also selective against the AT1 receptor and cell active.
| CID/ML# | Target Name | EC50 (nM) [SID, AID] | Anti-target Name(s) | EC50 (μM) [SID, AID] | Fold Selective |
|---|---|---|---|---|---|
| CID 46905036 ML233 | APJ Receptor | 3740 nM SID99361200 AID488985 | AT1 Receptor | >79 μM SID99361200 AID488986 | >21 |
Recommendations for scientific use of the probe
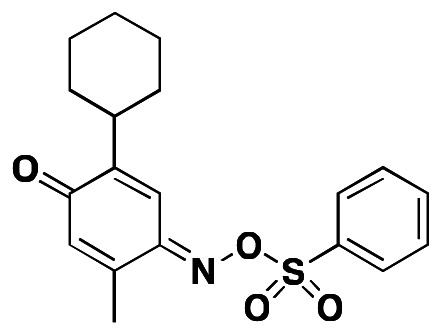
The recently discovered apelin system is emerging as a critical mediator of cardiovascular homeostasis (1,2) whose importance in the pathogenesis of hypertension (3–5), heart failure (6–8), atherosclerosis (9, 10) and other cardiovascular diseases is the subject of intense investigation (11). These investigations are limited by the paucity of research tools and reagents needed to fully understand the role of apelin in physiology and pathology. Thus a novel small molecule agonist of the apelin receptor (a.k.a. APJ, AGTRL-1, APLNR) is an essential tool for understanding the pharmacology of APJ and to validate the importance of this system in animal models of cardiovascular disease. Iturrioz et. al. (12) have reported E339-3D6 as the first non-peptide apelin receptor agonist. However, examination of the structure of E339-3D6 (see Fig. 1, below) weaken this claim as it is clearly a peptidomimetic of ~1400 m.w. Although E339-3D6 binds the apelin receptor competitively (Ki = 0.43 mM) it behaves as a partial agonist towards forskolin-induced cAMP production. Despite this moderate activity, E339-3D6 demonstrated some efficacy in vivo and in ex vivo tissues. To date, no small molecule has yet been identified as a full agonist. Disclosed in this report is the discovery and characterization of the first potent and selective apelin small molecule receptor agonist. Synthetic methodology, structure-activity relationships (SAR) and activity of our most potent and selective compound in a cell-based assay of apelin receptor activation are included. The probe molecule can be used as a tool compound for investigators interested in understanding apelin receptor pharmacology and function.
Figure 1
Reported “small molecule” APJ agonist.
1. Introduction
Currently there are no small molecule tools to investigate the biological functions of apelin and its receptor. Apelin is the endogenous peptide ligand for the G-protein coupled receptor (GPCR) APJ (angiotensin II receptor-like 1, AGTRL-1 and APLNR). Until the discovery of apelin, APJ was an orphan GPCR (13, 14). APJ is coupled to Gαi, and has been shown in cell culture to inhibit adenylate cyclase (15). The APJ gene encodes a receptor that most closely resembles the angiotensin receptor AT1. However, the APJ receptor does not bind angiotensin II (13). Underscoring the emerging importance of the apelin/APJ system, recent studies have shown that apelin reduces the extent of atherosclerotic lesions in ApoE−/− mice, and opposes the development of abdominal aortic aneurysms (9). Additionally, work in Dr. Smith’s lab revealed that APJ forms a heterodimer with the Ang II receptor AT1, and that this complex facilitates antagonism of Ang II signaling by apelin (LHS personal communication). Despite these exciting results, there remains a multitude of unanswered questions regarding the role of apelin and APJ in normal physiology and the pathogenesis of cardiovascular disease.
An agonist or potentiators of APJ would therefore provide a novel research tool to evaluate the role of apelin in cardiovascular and metabolic disease pathology. The criteria established was to identify (1) a transient and reversible full or partial agonist with an EC50 ≤ 5 μM in a functional assay (cell based) and (2) selective for APJ over AT1 by at least 30-fold.
There has been one recent report of an APJ agonist in the literature, E339-3D6 (Figure 1). However the molecular weight exceeds 1000 Da (mw~1400) and the structure is clearly peptide related. Furthermore, E339-3D6 has a highly undesirable fluorescent probe attached to it, in addition to a urea, a methylated imidazole salt and a methylated aminothiazole salt. E336-3D6 is neither commercially available nor available from the authors. In addition to the report covering E339-3D6, several publications covering high molecular weight peptide derived APJ agonists have been reported in the patent literature (16–18).
2. Materials and Methods
The details of the primary HTS and additional assays can be found in the “Assay Description” section in the PubChem BioAssay view under the AIDs as listed in Table 1 below. Additionally the details for the primary HTS are provided in the Appendix at the end of this probe report.
Table 1
Summary of Assays and AIDs.
Primary HTS for APJ receptor agonism: Briefly, the primary assay detect agonists that cause the activation and internalization of the Angiotensin II receptor-like 1 (AGTRL-1; Apelin receptor; APJ) in a CHO-K1 AGTRL-1 Beta-Arrestin Cell Line (DiscoveRx) containing the stably integrated APJ receptor in reference to the canonical peptide APJ receptor agonist, the Apelin-13 peptide. Cells were seeded (1000 cells in 4 μL) in a 1536 well plate, grown overnight, then treated with 20 μM of compounds in 1% final DMSO, incubated at room temperature in the dark, and luminescence read 90 mins after addition of test compounds (See details in the Appendix).
Counterscreen for Angiotensin 1 receptor agonism: Briefly, this assay is performed in a completely analogous manner to the primary APJ agonist assay, except for the use of the angiotensin receptor 1 (AGTR-1; ATR-1) containing CHO-KI-AGTR-1 cell line (DiscoveRx). This assay assesses the selectivity of APJ agonist against this closely related receptor to the APJ receptor. (See details in the Appendix)
2.1. Assays
Table 1 summarizes the details for the assays that drove this probe project.
2.2. Probe Chemical Characterization
a. Chemical name of probe compound
The IUPAC name of the probe is (E)-2-cyclohexyl-5-methyl-4-(phenylsulfonyloxyimino)cyclohexa-2,5-dienone. The specific batch prepared, tested and submitted to the MLSMR is archived as SID99361200 corresponding to CID46905036.
b. Probe chemical structure including stereochemistry if known
The probe ML233 was obtained as a single observed anti isomer. The assignment is consistent with precedent literature (19).
c. Synthesis and Structural Verification Information of probe SID99361200 corresponding to CID46905036

Scheme 1Synthesis of ML233, conditions
a. Sodium nitrite (1.5 eq.), HCl (conc.), EtOH; b. benzenesulfonyl chloride (1 eq.), DMAP (cat.), pyridine.
For detailed procedures see Section 2.3. Images of spectral data (1HNMR, 13CNMR, and LC/MS) used to support the structural assignment of ML233 can be found in Section 6 (Supplementary Information).
d. If available from a vendor, please provide details
This probe is not commercially available. A 25 mg sample of ML233 synthesized at SBCCG has been deposited in the MLSMR (see Probe Submission Table 3 below).
Table 3
Probe and Analog Submissions to MLSMR (BioFocus DPI) for APJ Agonists.
e. Solubility and Stability of probe in PBS at room temperature
The stability and solubility of ML233 was investigated in PBS buffer at room temperature (Figure 2). Initial experiments examining the stability of ML233 revealed no quantifiable level of compound after 1 hr in PBS buffer due to low solubility. Subsequent examination in 50% acetonitrile:PBS (1:1) showed the compound was mostly stable out to 48 hr (71% remaining).
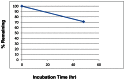
Figure 2
Stability of ML233 in PBS.
f. A tabulation of calculated and known probe properties
Table 2ML233 (CID46905036)
| Molecular Weight | 359.4393 [g/mol] |
| Molecular Formula | C19H21NO4S |
| AlogP | 4.697 |
| H-Bond Donor | 0 |
| H-Bond Acceptor | 5 |
| Rotatable Bond Count | 4 |
| Exact Mass | 359.43933 |
| MonoIsotopic Mass | 359.43933 |
| Topological Polar Surface Area | 81.18 |
| Heavy Atom Count | 25 |
| Formal Charge | 0 |
| Complexity | 646 |
| Isotope Atom Count | 0 |
| Defined Atom StereoCenter Count | 0 |
| Undefined Atom StereoCenter Count | 0 |
| Defined Bond StereoCenter Count | 1 |
| Undefined Bond StereoCenter Count | 0 |
| Covalently-Bonded Unit Count | 1 |
g. Provide MLS# that verifies the submission of probe molecule and five related samples that were submitted to the SMR collection
Samples of the probe (25 mg) and each of five analogs (20 mg) synthesized at SBCCG were submitted to MLSMR (Table 3), and 5 mg of the probe was provided to Dr. Smith.
2.3. Probe Preparation
Step 1: 2-cyclohexyl-5-methylphenol (0.5g, 2.63mmol) was dissolved in 10 ml ethanol followed by addition of 10 ml concentrated HCl. The resultant mixture was cooled to 0 °C and sodium nitrite (272mg, 3.94mmol) was added to it in two portions of about 140mg each. After addition of sodium nitrite, the reaction mixture turned green. The reaction mixture was stirred overnight at room temperature followed by addition of ice-cold water, which resulted in precipitation of a yellow solid. The aqueous solution was filtered under vacuum to yield 0.55g (96% yield) of 2-cyclohexyl-5-methyl-4-nitrosophenol (2) as a yellow solid. 1H NMR (500 MHz, CDCl3) δ 8.69 (s, 1H), 7.47 (s, 1H), 6.30 (d, J = 1.2 Hz, 1H), 2.74 (t, J = 12.0 Hz, 1H), 2.15 (d, J = 1.1 Hz, 3H), 1.88 – 1.65 (m, 5H), 1.49 – 1.31 (m, 2H), 1.29 – 1.07 (m, 3H).
Step 2: 2-cyclohexyl-5-methyl-4-nitrosophenol (50mg, 0.23mmol) was dissolved in 2ml pyridine followed by addition of catalytic amount of DMAP (N, N′-Dimethylaminopyridine). Benzenesulfonyl chloride (40mg, 0.23mmol) was added to the reaction mixture and the reaction mixture was stirred for 4–6h. The reaction mixture was partitioned with approximately 20 ml of 1:1 ethyl acetate and water and the organic layer was washed with 10% HCl (2×20ml) and brine (1×20ml). The organic layer was concentrated in vacuo and the crude product was purified by column chromatography to yield 60mg (73%) of 4-methyl-5-(((phenylsulfonyl)oxy)imino)-[1,1′-bi(cyclohexane)]-3,6-dien-2-one as a yellow solid. 1H NMR (500 MHz, CDCl3) δ 8.12 – 7.99 (m, 2H), 7.73 (t, J = 7.5 Hz, 1H), 7.62 (t, J = 7.8 Hz, 2H), 7.28 (d, J = 0.8 Hz, 1H), 6.33 (d, J = 1.3 Hz, 1H), 2.75 (dt, J = 21.2, 7.4 Hz, 1H), 2.10 (d, J = 1.3 Hz, 3H), 1.92 – 1.69 (m, 5H), 1.47 – 1.31 (m, 2H), 1.32 – 1.12 (m, 3H). 13C NMR (125 MHz, CDCl3) δ 185.66, 154.32, 151.39, 144.11, 134.74, 134.60, 131.50, 129.23, 129.11, 118.61, 36.57, 32.29, 26.37, 26.01, 16.77.
3. Results
3.1. Summary of Screening Results
The flowchart on the right summarizes the screening results (Figure 3). Following primary HTS of approximately 330,000 Molecular Libraries Small Molecules Repository (MLSMR) compounds at 20 μM against the APJ receptor (AID2520), 347 initial actives (~0.1% hit rate) were obtained at a 30% activity cut-off which corresponded to a Z-score of 6.0. For a cell-based assay, the screen was robust with an average Z′ of 0.56.
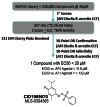
Figure 3
Flowchart Summary of Screening Results.
Fresh stock solution “cherry picks” of the 347 initial hits were requested from the MLSMR and 311 were received (89.6%) and tested (AID2764) and activities (EC50) were confirmed in full 10-point dose-response titrations (AID488748). In parallel, the selectivity against the angiotensin receptor 1 (ATR1) was also obtained (AID488865).
Only one tractable and reasonable compound/scaffold (quinone-oxime sulfonate) was obtained, that appeared selective against the AT1 receptor. However the potency (11.8 μM EC50 from liquid reorder) did not meet the desired 5 μM potency, and the level of agonism of the most potent compounds were in the 30 – 60% efficacy range. Interestingly, this compound was listed in PubChem with undefined stereochemistry. Upon reorder of the dry powder, the compound was found to be a single isomer CID5601088 (MLS-0437377), although reordered CID1909670 (MLS-0304385) had the same spectral properties and biological activity.
Over 40 additional analogs were available for purchase, which helped to guide the SAR and synthesis plan as detailed in the SAR sections below.
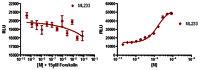
3.2. Dose Response Curves for Probe

Figure 4Dose response curves for ML233
3.3. Scaffold/Moiety Chemical Liabilities
Unlike the related quinone imine sulfonate scaffold, which is generally reactive and appears as a frequent hitter in bioassays, the quinone oxime sulfonate is stable and does not appear as a promiscuous compound from a Scifinder® search performed March 22, 2011.
To address concerns of potential hydrolytic stability or reactivity of ML233, an aliquot of the compound was prepared as a solution in 50% aqueous acetonitrile and was analyzed by LC/MS. A comparison at time zero, six, fifteen and thirty hours indicates the oxime and sulfonamide moiety is stable in aqueous solution (see Supplementary Information). In addition, an aliquot of probe ML233 was treated with a solution of glutathione to evaluate potential reactivity as an electrophile. The results indicate ML233 to be unreactive to glutathione over a seven-hour period (see Supplementary Information).
3.4. SAR Tables
The general SAR strategy we pursued around the quinone oxime sulfonate scaffold is depicted in Figure 5. The structure represented by MLS-0304385 (as noted above, upon reorder of the dry powder, the compound was found to be a single isomer CID5601088 (MLS-0437377)) emerged as the only scaffold from the HTS to demonstrate selectivity over the AT-1 receptor, which was a key element of the desired probe criteria. Once hydrolytic stability and lack of reactivity towards GSH were confirmed (see section 3.3), analog synthesis was initiated to develop SAR with a primary focus on aliphatic substitutions on the core quinone moiety (in green) and a range of substituted aryl sulfonates (in blue). The results are summarized in the table below (Table 4).

Figure 5
General SAR strategy around quinone-oxime sulfonate scaffold.
Table 4
SAR Analysis of APJ Agonists: Quinone-oxime sulfonates.
For the core quinone portion it was found that the optimal substitution consisted of a single larger aliphatic (for example isopropyl, t-butyl or cyclohexyl) flanking the oxygen (R1) combined with a smaller aliphatic group (such as methyl) flanking the imine (R3). For example, one of the most potent full agonists was achieved with R1= cyclohexyl and R3= methyl (Entry 6, ML233, CID46905036). Other combinations with R3=methyl that were potent were R1=isopropyl (Entries 5, 8, 10, 11, 13, 15 28–30) and R1=t-butyl (Entry 7). Interestingly, the reverse substitution with R3= isopropyl and R1= methyl gave less potent (Entry 18) or partial agonists (Entries 16–17).
In general, singly substituted analogs were inactive (Entries 21–27 and 40–42). Other combinations gave compounds that were either less active or showed only partial agonism. For example, with R1 and R2= methyl, only partial agonists were obtained (Entries 1–4), as with combinations of R1 and R3= methyl (Entries 9 and 12).
A range of substitution was tolerated on the pendant phenyl ring, including chloro, bromo, cyano, methyl and methoxy groups (Entries 32–39). Interestingly, a 3-pyridyl was found to be a suitable replacement for phenyl (Entry 31, compared to entry 11), providing opportunities to further improve the solubility of this series.
3.5. Cellular Activity
All of the primary and selectivity assays are cell based. Specific measures of permeability and toxicity are discussed in section 3.6 below.
3.6. Profiling Assays
The nominated probe was evaluated in a detailed in vitro pharmacology screen as shown in Table 5.
Table 5
Summary of in vitro ADME Properties of APJ Antagonist probe ML233.
ML233 is poorly soluble in aqueous media at pH 5.0/6.2/7.4.
The PAMPA (Parallel Artificial Membrane Permeability Assay) assay is used as an in vitro model of passive, transcellular permeability. An artificial membrane immobilized on a filter is placed between a donor and acceptor compartment. At the start of the test, drug is introduced in the donor compartment. Following the permeation period, the concentration of drug in the donor and acceptor compartments is measured using UV spectroscopy. Consistent with the predicted LogP (see Table 3), ML233 is a highly permeable compound in this assay. This data suggests that, like many therapeutic molecules, the poor aqueous solubility of ML233 will have little impact on its activity in cells and tissues.
Plasma Protein Binding is a measure of a drug’s efficiency to bind to the proteins within blood plasma. The less bound a drug is, the more efficiently it can traverse cell membranes or diffuse. Highly plasma protein bound drugs are confined to the vascular space, thereby having a relatively low volume of distribution. In contrast, drugs that remain largely unbound in plasma are generally available for distribution to other organs and tissues. ML233 is highly bound to plasma proteins in mouse plasma (99% bound). Similarly, although to a lesser extent, ML233 is highly protein bound in human plasma (98% bound). This interaction may confer stability to the molecule in plasma, protecting it from metabolizing enzymes.
Plasma Stability is a measure of the stability of small molecules and peptides in plasma and is an important parameter, which can strongly influence the in vivo efficacy of a test compound. Drug candidates are exposed in plasma to enzymatic processes (proteinases, esterases), and they can undergo intramolecular rearrangement or bind irreversibly (covalently) to proteins. Although ML233 is extensively bound to plasma proteins, the compound exhibits very poor stability in human plasma (18.68% remaining) after 3 hr. In contrast, ML233 is very stable in mouse plasma (100% remaining). This may be explained by the relative activities of metabolizing enzymes in the differing plasmas.
Alternatively, these disparate results may be explained by the subtle differences in plasma protein binding in mouse versus human plasma. Although the difference in percent compound bound in mouse versus human plasma appears minor (99% vs. 98%), the plasma protein-binding assay is not performed in a way that is suitable for assessing the kinetics of ML233 binding to plasma proteins. Therefore, we cannot exclude the possibility that as ML233 dissociates from human plasma proteins, it is rapidly metabolized.
The microsomal stability assay is commonly used to rank compounds according to their metabolic stability. This assay addresses the pharmacologic question of how long the parent compound will remain circulating in plasma within the body. ML233 shows poor stability (<1.0% remaining after 60 minutes) in both human and mouse liver homogenates. Ultimately this limits the utility of this probe to in vitro studies or apelin receptor or in vivo studies using acute intravenous doses to avoid hepatic metabolism.
ML233 shows some toxicity (LC50 = 25.8 μM) toward human hepatocyctes.
Profiling against other GPCRs. The probe, ML233 (CID46905036), was submitted to the Psychoactive Drug Screening Program (PDSP) at the University of North Carolina (PDSP, Bryan Roth, PI) and the data against a GPCR binding assay panel is shown in Figure 6. ML233 shows potentially significant binding against several other receptors, including the 5-HT1A, α2C adrenergic, and benzylpiperazine receptors (55%I, 51%I and 65%I at 10 μM, respectively) and norepinephrine transporter (57%I at 10 μM). It is not known whether these activities in binding assays are translated into functional modification of the activities of these receptors. In addition, these potential cross-reactivities should not confound any in vitro data obtained with ML233.

Figure 6
GPCR profiling panel for ML233.
As a follow up experiment, ML233 was submitted for further testing by DiscoveRx (as a CRO) in a panel of functional assays of selected GPCRs (see Table 6 below). Those GPCRs tested were chosen based on their logical association with apelin/APJ due to their effects on cardiovascular function. Table 6 shows that ML233 has little to no agonist activity at 14 related receptors when assayed at 10μM. Those receptors tested include the adrenergic receptor family, the endothelin and bradykinin receptor families, as well as the vasopressin receptors. In this panel of GPCRs, the AT1 receptor was also profiled (AGTR1, line 7). The data returned from this externally executed panel is shown below in Table 6.
Table 6
ML233 Functional profiling data against the DiscoveRx GPCR Panel tested at a single 10 μM concentration.
4. Discussion
4.1. Comparison to existing art and how the new probe is an improvement
There has been only one report of a “small molecule” APJ agonists in the open or patent literature (12), but, as discussed in the introduction, this is a peptide derived compound with mw>1000. Thus, ML233 represents the first true small molecule (mw= 359) APJ agonist that will be available to the scientific community.
4.2. Mechanism of Action Studies
Dr. Smith’s laboratory has performed a series of exploratory studies to investigate the mechanism of action of this probe. The mechanism of action of ML233 is that of a classic full agonist of APJ in the cell-based assays employed in this project. Because APJ is capable of both G-protein-dependent and β-arrestin-dependent signaling, and the primary assay utilized in the project measured the activity of ML233 as a function of β-arrestin recruitment to APJ, we assessed the activity of ML233 in an assay of G-protein signaling by APJ. This MOA assay uses the High Sensitivity Lance TR-FRET cAMP assay (Perkin Elmer). Consistent with its role as an APJ agonist, ML233 reduced forskolin stimulated increases in intracellular cAMP (Figure 7A). Interestingly, the observed potency of ML233 to reduce cAMP was significantly higher than the maximal concentration studied (100μM). In addition, ML233 induced APJ internalization (Figure 7B) taken together these probe characterization studies suggest that, like most GPCRs, the apelin receptor is capable of pleiotropic signaling, and that ML233 is a useful tool probe β-arrestin-mediated signaling and receptor internalization of APJ. Those investigators specifically interested in these aspects of APJ pharmacology and function will be well served by using ML233. However, the utility of ML233 in probing G-protein signaling of APJ is yet to be fully explored. Investigators using ML233 for this purpose should further explore the effects of ML233 in their assay of choice.
4.3. Planned Future Studies
As described above, studies to support the proposed mechanism of action are underway in Dr. Smith’s laboratory using the probe molecule described in this report. Future studies also planned will involve testing ML233 in genetically tractable model organisms including zebra fish and mice. Additional medicinal chemistry efforts to improve the drug-like properties of ML233 are under way. Specific efforts will be directed at improving the metabolic stability and especially solubility. Improvements in these properties will improve the in vivo utility of ML233 in vivo.
Future in vitro studies to be performed by Dr. Smith will determine if ML233 is a direct competitor of apelin binding using standard competition binding studies and radiolabeled apelin-13. Consistent with our Chemical Probe Development Plan, we will also evaluate the affinity of ML233 to the angiotensin II type 1 receptor, the receptor most closely related to APJ. Additional studies will utilize existing cell based assays to determine if ML233 is an allosteric modulator of APJ
5. References
- 1.
- Quazi R, Palaniswamy C, Frishman WH. The emerging role of apelin in cardiovascular disease and health. Cardiol Rev. 2009;17:283–286. [PubMed: 19829178]
- 2.
- Sorli SC, van den Berghe L, Masri B, Knibiehler B, Audigier Y. Therapeutic potential of interfering with apelin signalling. Drug Discov Today. 2006;11:1100–1106. [PubMed: 17129829]
- 3.
- Chandra SM, Razavi H, Kim J, Agrawal R, Kundu RK, de Jesus Perez V, Zamanian RT, Quertermous T, Chun HJ. Disruption of the Apelin-APJ System Worsens Hypoxia-Induced Pulmonary Hypertension. Arterioscler Thromb Vasc Biol. 2010;31:814–820. [PMC free article: PMC3113525] [PubMed: 21233449]
- 4.
- Niu W, Wu S, Zhang Y, Li W, Ji K, Gao P, Zhu D. Validation of genetic association in apelin-AGTRL1 system with hypertension in a larger Han Chinese population. J Hypertens. 2010;28:1854–1861. [PubMed: 20485192]
- 5.
- Przewlocka-Kosmala M, Kotwica T, Mysiak A, Kosmala W. Reduced circulating apelin in essential hypertension and its association with cardiac dysfunction. J Hypertens. 2011 [PubMed: 21346619]
- 6.
- Ashley EA, Powers J, Chen M, Kundu R, Finsterbach T, Caffarelli A, Deng A, Eichhorn J, Mahajan R, Agrawal R, et al. The endogenous peptide apelin potently improves cardiac contractility and reduces cardiac loading in vivo. Cardiovasc Res. 2005;65:73–82. [PMC free article: PMC2517138] [PubMed: 15621035]
- 7.
- Chen MM, Ashley EA, Deng DX, Tsalenko A, Deng A, Tabibiazar R, Ben-Dor A, Fenster B, Yang E, King JY, et al. Novel role for the potent endogenous inotrope apelin in human cardiac dysfunction. Circulation. 2003;108:1432–1439. [PubMed: 12963638]
- 8.
- Jia YX, Pan CS, Zhang J, Geng B, Zhao J, Gerns H, Yang J, Chang JK, Tang CS, Qi YF. Apelin protects myocardial injury induced by isoproterenol in rats. Regul Pept. 2006;133:147–154. [PubMed: 16278022]
- 9.
- Chun HJ, Ali ZA, Kojima Y, Kundu RK, Sheikh AY, Agrawal R, Zheng L, Leeper NJ, Pearl NE, Patterson AJ, et al. Apelin signaling antagonizes Ang II effects in mouse models of atherosclerosis. J Clin Invest. 2008;118:3343–3354. [PMC free article: PMC2525695] [PubMed: 18769630]
- 10.
- Hashimoto T, Kihara M, Imai N, Yoshida S, Shimoyamada H, Yasuzaki H, Ishida J, Toya Y, Kiuchi Y, Hirawa N, et al. Requirement of apelin-apelin receptor system for oxidative stress-linked atherosclerosis. Am J Pathol. 2007;171:1705–1712. [PMC free article: PMC2043530] [PubMed: 17884970]
- 11.
- Barnes G, Japp AG, Newby DE. Translational promise of the apelin--APJ system. Heart. 2010;96:1011–1016. [PubMed: 20584856]
- 12.
- Iturrioz X, Alvear-Perez R, De Mota N, Franchet C, Guillier F, Leroux V, Dabire H, Le Jouan M, Chabane H, Gerbier R, et al. Identification and pharmacological properties of E339-3D6, the first nonpeptidic apelin receptor agonist. FASEB J. 2010;24:1506–1517. [PubMed: 20040517]
- 13.
- O’Dowd BF, Heiber M, Chan A, Heng HH, Tsui LC, Kennedy JL, Shi X, Petronis A, George SR, Nguyen T. A human gene that shows identity with the gene encoding the angiotensin receptor is located on chromosome 11. Gene. 1993;136:355–360. [PubMed: 8294032]
- 14.
- Tatemoto K, Hosoya M, Habata Y, Fujii R, Kakegawa T, Zou MX, Kawamata Y, Fukusumi S, Hinuma S, Kitada C, et al. Isolation and characterization of a novel endogenous peptide ligand for the human APJ receptor. Biochem Biophys Res Commun. 1998;251:471–476. [PubMed: 9792798]
- 15.
- Hosoya M, Kawamata Y, Fukusumi S, Fujii R, Habata Y, Hinuma S, Kitada C, Honda S, Kurokawa T, Onda H, et al. Molecular and functional characteristics of APJ. Tissue distribution of mRNA and interaction with the endogenous ligand apelin. J Biol Chem. 2000;275:21061–21067. [PubMed: 10777510]
- 16.
- Janz J, Kuliopulos A, McMurry TJ. WO2010/053545. APJ Receptor Compounds.
- 17.
- Llorens-Cortes C, Guillier F, Franchet C. EP1903052. APJ receptor ligands and uses thereof. 2006
- 18.
- Hamada J, Kimura J, Ishida J, Kohda T, Morishita S, Ichihara S, Fukamizu A. Evaluation of novel cyclic analogues of apelin. International Journal of Molecular Medicine. 2008;22:547–552. [PubMed: 18813863]
- 19.
- Ishikawa T, Saito T, Ishii H. Synthesis of Macarpine and its Cytotoxicity: Toward a Synthetic Route for 12-Alkoxybenzo[c]phenanthridine Alkaloids through Aromatic Nitrosation under Basic Condition. Tetrahedron. 1995;51:8447–8458.
6. Supplementary Information
6.1. Assay Details: APJ Beta-Arrestin 1536-Well Agonist Assay Protocol
A. Brief Description of the Assay
The purpose of this assay is to detect agonists that cause the activation and internalization of the Angiotensin II receptor-like 1 (Apelin receptor) in the CHO-K1 AGTRL-1 Beta-Arrestin Cell Line in 1536-well plate format.
B. Materials
- Angiotensin II receptor-like 1 (AGTRL-1) Cell Line (DiscoveRx, Cat# 93-0250C2)
- F12 nutrient mix HAMs (Invitrogen, Cat# 11765)
- Fetal Bovine Serum, heat-inactivated (Hyclone, Cat# SH30396)
- 100X Penicillin/Streptomycin Solution (Invitrogen, Cat#15140-122)
- Hygromycin B (Roche, Cat# 10843555001)
- Geneticin (MPBiomedicals, Cat # 1672548)
- Trypsin-EDTA 0.25% (Invitrogen, Cat# 25200-056)
- Cell Dissociation Buffer (Invitrogen, Cat # 13151)
- DPBS (Hyclone, Cat# 30028.02)
- T225 TC Flask (Nunc, Cat# 159934)
- Cell strainer, 40 um (BD, Cat# 352340)
- 1536-well, white, solid-bottom, Kalypsys compatible, TC plate (Corning)
- Apelin-13 (Sigma-Aldrich, Cat # A6469)
- PathHunter Detection Reagents (DiscoveRx, Cat# 93-0001)
- Galacton Star
- Emerald 11
- Cell Assay Buffer
C. Procedures
Day1 –Cell Seeding
- Plate 1000 cells/well in 4 μL of assay media into columns 1–48 of a 1536-well assay plate, using straight tip dispense on a Kalypsys dispenser.
- Centrifuge plates at 500 rpm for 1 minute on a Vspin centrifuge. Use Kalypsys metal lids.
- Incubate overnight at 37 degrees, 100% relative humidity, 5% CO2 for 16–18 hours.
Day2 –Compound Addition
- 4.
Using the Kalypsys Dispenser, add 2μl/well of Assay media to Col. 3–48 for the negative control and test compound wells.
- 5.
Centrifuge plates at 500 rpm for 1 minute on a Vspin centrifuge.
- 4.
Using LabCyte Echo, transfer appropriate volume of test compounds in DMSO into assay plate Col. 5 – 48, then backfill with DMSO to equalize DMSO concentration if necessary (Final concentration primary assay= 1.0% DMSO, Final concentration dose response= 1.32%).
- 6.
Transfer equal volume of DMSO to positive and negative control wells in Columns 1 – 4.
- 7.
Immediately following compound/DMSO transfer via the Echo, transfer 2μl/well of 6 μM Apelin-13 in assay media to the positive control wells.
- 8.
Centrifuge plates at 500 rpm for 1 minute on a Vspin centrifuge.
- 9.
Incubate plates at room temperature in the dark for 90 minutes.
- 10.
Following 90 minute incubation, deliver 3.0 μL of Detection Reagent solution to each assay plate (Columns 1 – 48) using a Kalypsys dispenser.
- 11.
Centrifuge plates at 1000 rpm for 1 minute on a Vspin centrifuge.
- 12.
Incubate plates for 60 minutes at room temperature in the dark.
- 13.
Read plates using a Perkin Elmer Envision using a luminescence
D. Recipe
- Growth Media: F12 nutrient mix HAMs supplemented with 10% hi-FBS, 1X Penicillin/Streptomycin, 300μg/ml Hygromycin B, 800μg/ml Geneticin
- Assay Media: Same as Growth Media
- Trypsin: Dilute 0.25% Trypsin/EDTA to 0.05% Trypsin/EDTA using DPBS
- Positive Control: Assay Media with Apelin-13 (10 nM FAC)
- Negative Control: Assay Media with no Apelin-13
- Detection Reagent: Use the following ratio to prepare the detection reagent: Galacton Star: Emerald: Cell Assay Buffer (1:5:19)
6.2. Assay Details: AT1 Beta-Arrestin 1536-Well Agonist Assay Protocol
A. Brief Description of the Assay
The purpose of this assay is to detect agonists that cause the activation and internalization of the Angiotensin II receptor type 1 (AT1 receptor) in the CHO-K1 AGTR-1 Beta-Arrestin Cell Line in 1536-well plate format.
B. Materials
- Angiotensin II receptor type 1 (AGTRL-1) Cell Line (DiscoveRx, Cat# 93-0312C2)
- F12 nutrient mix HAMs (Invitrogen, Cat# 11765)
- Fetal Bovine Serum, heat-inactivated (Hyclone, Cat# SH30396)
- 100X Penicillin/Streptomycin Solution (Invitrogen, Cat#15140-122)
- Hygromycin B (Roche, Cat# 10843555001)
- Geneticin (MPBiomedicals, Cat # 1672548)
- Trypsin-EDTA 0.25% (Invitrogen, Cat# 25200-056)
- Cell Dissociation Buffer (Invitrogen, Cat # 13151)
- DPBS (Hyclone, Cat# 30028.02)
- T225 TC Flask (Nunc, Cat# 159934)
- Cell strainer, 40 μm (BD, Cat# 352340)
- 1536-well, white, solid-bottom, Kalypsys compatible, TC plate (Corning)
- Apelin-13 (Sigma-Aldrich, Cat # A6469)
- PathHunter Detection Reagents (DiscoveRx, Cat# 93-0001)
- Galacton Star
- Emerald 11
- Cell Assay Buffer
C. Procedures
Day1 –Cell Seeding
- Plate 500 cells/well in 5 μL of assay media into columns 1–48 of a 1536-well assay plate, using straight tip dispense on a Kalypsys dispenser.
- Centrifuge plates at 500 rpm for 1 minute on a Vspin centrifuge. Use Kalypsys metal lids.
- Incubate overnight at 37 degrees, 100% relative humidity, 5% CO2 for 16–18 hours.
Day2 –Compound Addition
- 5.
Using the Kalypsys Dispenser, add 1 μl/well of Assay media to Col. 3–48 for the negative control and test compound wells.
- 6.
Centrifuge plates at 500 rpm for 1 minute on a Vspin centrifuge.
- 7.
Using LabCyte Echo, transfer appropriate volume of test compounds in DMSO into assay plate Col. 5 – 48, then backfill with DMSO to equalize DMSO concentration (Final concentration = 1.32% DMSO).
- 8.
Transfer equal volume of DMSO to positive and negative control wells in Columns 1 – 4.
- 9.
Immediately following compound/DMSO transfer via the Echo, transfer 1μl/well of 6 μM Angiotensin II in assay media to the positive control wells.
- 10.
Centrifuge plates at 500 rpm for 1 minute on a Vspin centrifuge.
- 11.
Incubate plates at room temperature in the dark for 90 minutes.
- 12.
Following 90 minute incubation, deliver 3.0 μL of Detection Reagent solution to each assay plate (Columns 1 – 48) using a Kalypsys dispenser.
- 13.
Centrifuge plates at 1000 rpm for 1 minute on a Vspin centrifuge.
- 14.
Incubate plates for 60 minutes at room temperature in the dark.
- 15.
Read plates using a Perkin Elmer Envision using a luminescence
D. Recipe
- Growth Media: F12 nutrient mix HAMs supplemented with 10% hi-FBS, 1X Penicillin/Streptomycin, 300μg/ml Hygromycin B, 800μg/ml Geneticin
- Assay Media: Same as Growth Media except 6% hi-FBS
- Trypsin: Dilute 0.25% Trypsin/EDTA to 0.05% Trypsin/EDTA using DPBS
- Positive Control: Assay Media with Angiotensin II (FAC = 1 μM)
- Negative Control: Assay Media with no Angiotensin II
- Detection Reagent: Use the following ratio to prepare the detection reagent: Galacton Star: Emerald: Cell Assay Buffer (1:5:19)
- PMCPubMed Central citations
- PubChem BioAssay for Chemical ProbePubChem BioAssay records reporting screening data for the development of the chemical probe(s) described in this book chapter
- PubChem SubstanceRelated PubChem Substances
- PubMedLinks to PubMed
- Review Functional antagonists of the Apelin (APJ) receptor.[Probe Reports from the NIH Mol...]Review Functional antagonists of the Apelin (APJ) receptor.Maloney PR, Khan P, Hedrick M, Gosalia P, Milewski M, Li L, Roth GP, Sergienko E, Suyama E, Sugarman E, et al. Probe Reports from the NIH Molecular Libraries Program. 2010
- Discovery of 4-oxo-6-((pyrimidin-2-ylthio)methyl)-4H-pyran-3-yl 4-nitrobenzoate (ML221) as a functional antagonist of the apelin (APJ) receptor.[Bioorg Med Chem Lett. 2012]Discovery of 4-oxo-6-((pyrimidin-2-ylthio)methyl)-4H-pyran-3-yl 4-nitrobenzoate (ML221) as a functional antagonist of the apelin (APJ) receptor.Maloney PR, Khan P, Hedrick M, Gosalia P, Milewski M, Li L, Roth GP, Sergienko E, Suyama E, Sugarman E, et al. Bioorg Med Chem Lett. 2012 Nov 1; 22(21):6656-60. Epub 2012 Sep 7.
- Discovery of a novel small molecule agonist scaffold for the APJ receptor.[Bioorg Med Chem. 2016]Discovery of a novel small molecule agonist scaffold for the APJ receptor.Narayanan S, Maitra R, Deschamps JR, Bortoff K, Thomas JB, Zhang Y, Warner K, Vasukuttan V, Decker A, Runyon SP. Bioorg Med Chem. 2016 Aug 15; 24(16):3758-70. Epub 2016 Jun 11.
- Review Apelin and its receptor APJ in cardiovascular diseases.[Clin Chim Acta. 2014]Review Apelin and its receptor APJ in cardiovascular diseases.Yu XH, Tang ZB, Liu LJ, Qian H, Tang SL, Zhang DW, Tian GP, Tang CK. Clin Chim Acta. 2014 Jan 20; 428:1-8. Epub 2013 Sep 18.
- Discovery and SAR of aryl hydroxy pyrimidinones as potent small molecule agonists of the GPCR APJ.[Bioorg Med Chem Lett. 2020]Discovery and SAR of aryl hydroxy pyrimidinones as potent small molecule agonists of the GPCR APJ.Myers MC, Bilder DM, Cavallaro CL, Chao HJ, Su S, Burford NT, Nayeem A, Wang T, Yan M, Langish RA, et al. Bioorg Med Chem Lett. 2020 Apr 1; 30(7):126955. Epub 2020 Jan 22.
- Functional Agonists of the Apelin (APJ) Receptor - Probe Reports from the NIH Mo...Functional Agonists of the Apelin (APJ) Receptor - Probe Reports from the NIH Molecular Libraries Program
Your browsing activity is empty.
Activity recording is turned off.
See more...