NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
Scavenger receptor, class B, type I (SR-BI) mediates selective uptake of cholesterol from high density lipoprotein (HDL) particles, a poorly understood process that is distinct from endocytic uptake of lipoproteins, such as low density lipoprotein (LDL). We set out to find small molecules that modulate SR-BI function and could be used to characterize the mechanisms involved in HDL uptake. Using a cell-based DiI-HDL uptake assay, we performed a high-throughput screen (HTS) of the National Institutes of Health Molecular Libraries Probe Centers Network (NIH MLPCN) compound library. Of 319,533 compounds, 3,046 compounds (0.96%) were classified as inhibitors of DiI-HDL uptake. A bis-amide (CID 650573) was identified in the primary HTS as an inhibitor. It had potent activity upon retesting in the primary assay, a low hit rate in other MLPCN assays, and possessed structural properties suitable for analog synthesis. Structure activity relationship (SAR) studies were performed to improve potency and to minimize deleterious properties. These efforts generated a probe (CID 53393835/ML279), a low nanomolar inhibitor with improved potency and decreased metabolic liability. ML279 was tested for efflux inhibition, modulation of HDL binding to SR-BI, and inhibition of endocytosis. ML279 functions by inhibiting both SR-BI-mediated uptake and efflux of free cholesterol to HDL particles. As a tool compound, ML279 is superior to existing small-molecule inhibitors of SR-BI (e.g., BLT-1 and ITX-5061) and will be useful in elucidating how SR-BI mediates lipid transport. In addition, it could clarify the role of SR-BI in a number of biological processes where it plays a crucial role.
Assigned Assay Grant No.: 2 R01 HL052212-11
Screening Center Name & PI: Broad Institute Probe Development Center, Stuart Schreiber
Chemistry Center Name & PI: Broad Institute Probe Development Center, Stuart Schreiber
Assay Submitter & Institution: Monty Krieger, Biology Department, Massachusetts Institute of Technology
PubChem Summary Bioassay Identifier (AID): 488952
Probe Structure and Characteristics
| CID/ML # | Target | IC50 (µM) [SID, AID] | Antitarget | IC50 (μM) [SID, AID] | Fold Selective | Secondary Assays IC50 (µM) [SID, AID] |
|---|---|---|---|---|---|---|
| CID 53393835/ML279 | mSR-BI- mediated HDL uptake | IC50= 0.016 [SID 125333454, AID 588828] | 24-hour cytotoxicity | >35 [SID 125333454, AID 588829] | >1000x | 1) 3 hour cytotoxicity >35 [SID 115950079, AID 588830] 2) [ldlA7] DiI-HDL uptake >35 [SID 125333454, AID 588825] |
Recommendations for scientific use of the probe
The end goal of this project is to identify and to use small molecules to analyze the mechanisms of the HDL receptor, which is specifically referred to as the Scavenger receptor class B, type I (SR-BI) (1). SR-BI was the first high-density lipoprotein (HDL, ‘good’ cholesterol) receptor identified. SR-BI plays multiple roles in mammalian development, cellular physiology, and pathophysiology. It controls the structure and composition of plasma HDL, and levels and fates of HDL cholesterol, including delivery to the liver and steroidogenic tissues. SR-BI binds HDL and functions as a cell surface transporter to move cholesterol or its esters into or out of cells and as a signaling receptor to control cell function. SR-BI can also interact with and transport a plethora of other ligands. A new probe could be used to map important sites of interaction on SR-BI for these processes, to help identify possible intracellular binding partners, to verify whether SR-BI oligomerizes to mediate HDL interactions, or to improve our knowledge of other aspects of SR-BI biology.
We set out to identify probes with minimal cell cytotoxicity and fewer off-target effects compared with current SR-BI inhibitors. Blocker of Lipid Transport-1 (BLT-1) and several other inhibitors were identified in a pilot small molecule screen as an inhibitor of lipid uptake, with nanomolar potency in the case of BLT-1 (2). BLT-1 has been very useful in characterizing some aspects of SR-BI's role in lipid uptake (3,4,5). Although BLT-1 has been shown to have nanomolar potency, it binds SR-BI irreversibly and is extremely toxic to cells, which has limited the use of the compound to short term in vitro assays.
The probe (ML279), which is described in this report, is a reversible binder without related cytotoxicity and has similar measured potency relative to BLT-1 (see Section 4.1), despite being a reversible inhibitor. Another molecule described in the literature, ITX-5061, disrupts HDL uptake but with lower potency than ML279. In addition, ITX-5061 was originally developed to inhibit p38 MAP kinase activity (6). ML279 has shown an IC50 value of 16 nM compared to 254 nM for ITX-5061 in the primary assay (AID 588828). To date, we have not investigated ML279 against any kinases. Testing ML279 for off-target activities, including kinases, will be a part of early follow up studies.
SR-BI has a wide variety of functions in physiology and pathophysiology. Lipoprotein metabolism impacts endothelial biology, platelet function, bile secretion, female fertility, steroidogenesis and cholesterol homeostasis (7). SR-BI is considered to be a pattern-recognition receptor (PRP), a type of immune recognition receptor for microbial substances, such as lipopolysaccharide (LPS; 8). SR-BI has the ability to clear LPS and suppress stimulation of NF-kB and cytokine stimulation via several Toll-like receptors (TLRs; 9,10). A probe that blocks SR-BI in the context of immune regulation could be useful in dampening the immune response, especially in the case of sepsis.
SR-BI serves as a co-receptor for Hepatitis C (HCV) viral entry, and blockade with compound or antibodies can reduce infection (11,12,13,14). It is possible that like ITX-5061, ML279 will block HCV infection. SR-BI also seems to enhance infection of liver cells by the malaria parasite, Plasmodium falciparum (15, 16). Blockade of SR-BI by small molecules, such as ML279, may help us to understand the precise mechanisms that viruses and pathogens use to enter human cells and cause disease.
1. Introduction
Scientific Rationale
Developing tools to analyze SR-BI function and mechanism of action, as well as the manipulation of the activity of SR-BI in vitro and in vivo, will have significant impact on our understanding in diverse areas of physiology and pathophysiology of considerable medical importance. Indeed, recent disappointments in attempts to develop HDL-focused pharmaceutical agents with other molecular targets highlight the importance of developing a deeper and broader understanding of all aspects of HDL metabolism, including those mediated by SR-BI. Given the central role of SR-BI in lipid transfer and metabolism, inhibitors of SR-BI function will be useful tools to further probe the mechanism of SR-BI, and may also have clinical utility(such as pathogen cell entry (e.g., HCV).
Several small-molecule inhibitors of SR-BI have been discovered (Figure 1). In 2002, Krieger and coworkers reported five compounds (BLTs 1-5) discovered via HTS that block lipid transport selectively via SR-BI, with nanomolar to micromolar potencies.(6) Unfortunately, these probes exhibit an unacceptable level of cellular toxicity. The actions of four of these BLT compounds were confirmed to be on SR-BI itself, using assays with purified SR-BI reconstituted into liposomes (17). The diabetes drug glyburide, a potent inhibitor of the sulfonylurea receptors SUR1 and SUR2, was also found to have activity at SR-BI, though it was a relatively weak inhibitor of cholesterol efflux (3). Researchers at Sankyo discovered that the protected piperazine R-138329 increased HDL-cholesterol in both mice and hamsters, presumably by inhibiting SR-BI-mediated uptake of HDL (18). Recently, a p38 MAP kinase inhibitor (ITX-5061), was reported to have similar in vivo effects and appears to be a moderate inhibitor of SR- BI-mediated cholesterol uptake (6). ITX-5061 is an inhibitor of HCV entry into cells, and is presently in Phase II trials as a HCV antiviral treatment. This compound was prepared in our laboratories and was profiled in our assays of interest. Finally, ITX-7650 has been described as an inhibitor of cholesterol uptake and HCV infection, but the structure remains undisclosed (12). See Section 4.1 for a further discussion of the prior art, as well as the search string table in Appendix G.
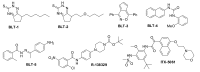
Figure 1
Reported Inhibitors of SR-BI. Inhibitors of SR-BI include the BLT compounds identified in a pilot screen, the piperazine and a kinase inhibitor.
A major goal of the proposed work is to identify a new SR-BI inhibitor that will provide a substantial improvement over the existing tool compounds under at least one of the following categories: 1) increased potency and reversibility; 2) reduced cellular toxicity; 3) new insights into mechanism; and 4) a unique activity profile (e.g., direct inhibition of ligand binding, differential influence on selective uptake, and free cholesterol efflux).
2. Materials and Methods
See subsections for a detailed description of the materials and methods used for each assay.
Materials and Reagents
- DiI-HDL, custom purified HDL particles derived from human blood were prepared by the Assay Provider and labeled with 1,1′-dioctadecyl-3,3,3′,3′- tetramethylindocarbocyanine perchlorate (DiI; Catalog No. D-282; Invitrogen, Carlsbad, CA).
- Alexa 488 HDL, human HDL particles labeled with the Alexa Fluor® 488 Protein Labeling Kit (Catalog No. A-10235, Invitrogen;Carlsbad, CA) were purified and labeled by the Assay Provider.
- CellTiter-Glo® Luminescent Cell Viability Assay was purchased from Promega (Catalog No. G7573; Madison, WI).
- Radiolabeled cholesterol [1,2-3H(N)]-, 1 mCi (37 MBq) was obtained from PerkinElmer – NEN (Catalog No. NET139001MC; Waltham, MA).
- Alexa Fluor-594 conjugated human transferrin (Catalog No. T-13343) was obtained from Invitrogen.
Cell Lines
The following cell lines were used in this study:
- ldlA[mSR-BI] is a Chinese hamster ovary (CHO) cell line that overexpresses SR-BI and was obtained from the Assay Provider (Krieger Laboratory). This cell line was used for the primary assay and several secondary assays.
- [ldlA7] is the parental cell line to ldlA[mSR-BI] cells, and does not overexpress SR-BI, and can be used to rule out compound activity independent of SR-BI. This cell line was obtained from the Assay Provider.
2.1. Assays
A summary listing of completed assays and corresponding PubChem AID numbers is provided in Appendix A (Table A1). Refer to Appendix B for the detailed assay protocols.
2.1.1. DiI-HDL Uptake Assay (Primary Assay AID 488896, AID 493194, AID 540354 AID 588392, AID 588553, AID 588548, AID 588754, AID 588828, AID 588833)
ldlA[mSR-BI] cells were plated into 384-well plates at 30 µl per well and incubated overnight. As a measurable surrogate for cholesterol uptake, human HDL particles were treated with the lipophilic fluorescent dye DiI and exposed to ldlA[mSR-BI] cells in lipoprotein-free media (Ham's F12/0.5% fatty acid-free Bovine Serum Albumin [BSA]/25 mM HEPES pH 7.4 plus 10 µg protein/ml DiI-HDL). Cells took up the Dil via SR-BI over 3 hours in the presence of compound. After significant uptake of the DiI, the cells became fluorescent. The level of fluorescence correlates with the amount of Dil uptake and can be measured with a standard plate reader. The uptake of lipid (represented by DiI) was inhibited by the compound BLT-1 or with an excess of HDL untreated with DiI.
The ldlA[mSR-BI] cells used in the assay were a CHO cell line lacking expression of the LDL receptors and overexpressing the Scavenger receptor (SR-BI). Inhibitors of SR-BI and HDL-mediated uptake lead to a reduction in fluorescence in this assay. Fluorescence was measured using a PerkinElmer EnVision plate reader. Primary HTS data was analyzed in Genedata Assay Analyzer, and were normalized against DMSO and 1 µM BLT-1 (positive control). For the HTS, the average of two replicates was used to rank order activity and to choose compounds for retests. For dose studies, the curves were generated with Genedata Condeseo and showed percent (%) activity for the individual doses.
2.1.2. Fluorescence Quencher Counterscreen (AID 588403, AID 540326)
The primary assay for this HDL project measures a reduction in fluorescence by potential inhibitors. One possible explanation is that the compounds have inherent fluorescence-quenching properties and reduce signal in a dose-dependent manner without actually altering any biology. We created an assay where compounds were pinned into assay buffer containing DiI-HDL without any cells. The compounds were incubated with DiI-HDL at 5 µg protein/ml in Ham's/25 mM HEPES/0.5% fatty acid-free BSA media for 30 minutes. Then, assay plates were measured with the identical settings for the primary assay. Compounds that quench DiI fluorescence lead to a dose-dependent loss of signal in this assay. Any compound that did not alter fluorescence or had an IC50 value of >30 µM was considered for further studies.
2.1.3. Cell Cytotoxicity Assay (AID 540246, AID 588829, AID 588826, AID 588830, AID 588842, AID 602126)
It is possible that a compound will cause cells to decrease HDL-mediated Dil uptake due to nonspecific consequences of cellular toxicity. Therefore, lead compounds should not be toxic in the 3-hour time period used in the primary assay. The cells were treated with compounds for 3 hours or 24 hours, and then cell viability was measured using the CellTiter-Glo Assay (Promega), which is a luciferase-based reagent that measures cellular ATP levels. The compounds were tested at different doses to determine IC50 values. Compounds that were toxic after 3 hours with an IC50 value of less than 30 µM were excluded from additional studies. Compounds that were active in the primary assay but toxic below 30 µM at 24 hours (but nontoxic at 3 hours) were still considered but none of our preferred scaffolds showed cytotoxicity at either timepoint (Figure 4C, 4D). Data were normalized against DMSO and BLT-1 (positive control) wells in Genedata Assay Analyzer. Curves were generated with Genedata Condeseo and showed percent (%) activity for the individual doses.
2.1.4. HDL Binding Assay (AID 588777, AID 588810)
HDL binding was assessed using Alexa Fluor 488-labeled HDL particles. For this assay, the Alexa 488 dye was directly covalently bound to apolipoprotein components of the HDL particle via primary amines; thus, no transfer of the fluorophore to cell membranes occurs. Instead, direct binding of the HDL particles to SR-BI can be measured. As a positive control, BLT-1 was used at 1 µM, which is known to increase binding (2). It is possible that a compound can reduce binding of HDL to the receptor, and this would lead to a decrease in signal. This assay is used to characterize the mechanism of action of a particular compound; therefore, any outcome in the assay is acceptable. Data were normalized against DMSO and BLT-1 (positive control) wells in Genedata Assay Analyzer. Curves were generated with Genedata Condeseo and showed percent (%) activity for the individual doses.
2.1.5. Cholesterol Efflux Assay (AID 588818, AID 602152)
SR-BI also mediates bidirectional flux (e.g., efflux) of unesterified or ‘free’ cholesterol (FC) between cells and HDL or other acceptors. This assay is used to determine if the compounds alter the efflux of free cholesterol to HDL. Compounds that do and do not inhibit in a dose-dependent manner will be of value.
On Day 0, 50,000 cells per well were plated in 24-well plates. On the following day, the media was removed and [3H]cholesterol in lipoprotein-deficient media was added. On Day 1, the medium was replaced with Ham's F12 medium supplemented with 10% bovine lipoprotein deficient serum, 1 µCi/ml [1,2-3H]cholesterol (40–60 Ci/mmol). On Day 3, the cells were washed to remove serum and incubated in Ham's F12 plus 1% fatty acid-free BSA. On Day 4, the cells were washed and pretreated with compounds for 1 hour. Subsequently, the cells were incubated for an additional 2 hours with the same concentrations of small molecules and with unlabeled HDL (final HDL concentration of 100 µg/ml). The media was collected to determine released cholesterol, and the cells were lysed. The amount of [3H]cholesterol in the media and cells was determined using liquid scintillation counting. Total cellular [3H]cholesterol was calculated as the sum of the radioactivity in the efflux medium plus the radioactivity in the cells and was used to calculate the [3H]cholesterol efflux (percent of total [3H]cholesterol released into the medium).
2.1.6. Mutant SR-BI Cholesterol Efflux Assay (AID 588843, AID 602138)
BLT-1 is known to interact at cysteine 384 of SR-BI (5). If the cysteine is mutated to serine, BLT-1 can no longer bind and shows no ability to inhibit cholesterol uptake. This assay is used to determine if the compounds work in a similar fashion to BLT-1 and require Cys384 to alter the efflux of FC to HDL. Compounds that do and do not inhibit efflux in a dose-dependent manner will be of value.
On Day 0, 50,000 C384S mutant cells per well were plated in 24-well plates. On the following day, the media was removed and [3H]cholesterol in lipoprotein-deficient media was added. On Day 1, the medium was replaced with Ham's F12 medium supplemented with 10% bovine lipoprotein deficient serum, 1 µCi/ml [1,2-3H]cholesterol (40–60 Ci/mmol). On Day 3, the cells were washed to remove serum and incubated in Ham's F12 plus 1% fatty acid-free BSA. On Day 4, the cells were washed and pretreated with compounds for 1 hour. Subsequently, the cells were incubated for an additional 2 hours with the same concentrations of small molecules and with unlabeled HDL (final HDL concentration of 100 µg protein/ml). The media was collected to determine released cholesterol, and the cells were lysed. The amount of [3H]cholesterol in the media and within cells was determined using liquid scintillation counting. Total cellular [3H]cholesterol was calculated as the sum of the radioactivity in the efflux medium plus the radioactivity in the cells and was used to calculate the [3H]cholesterol efflux (percent of total [3H]cholesterol released into the medium).
2.1.7. DiI-HDL Uptake Assay in the Absence of SR-BI (AID 588825)
In the primary assay, the cell line used to measure DiI-HDL uptake lacks the LDL receptor and overexpresses SR-B1. In this assay, the parental CHO cell line (ldlA7), which lacks the LDL receptor but does not overexpress SR-B1, was used to determine if inhibitors work via non-SR-BI-related mechanisms. As in the primary assay, 10 µg protein/ml of DiI-HDL was used to measure Dil uptake into the cells after 3 hours of incubation with different concentrations of compound. Little to no uptake of DiI-HDL was observed in these cells, and the only signal observed was minimal background staining.
2.1.8. Transferrin Endocytosis Assay (AID 602134)
This assay measures the endocytosis of an independent ligand, transferrin, which is not taken up via SR-BI but by clathrin-mediated endocytosis; thus providing a measure of the selectivity of the inhibitors. Alexa-594-labeled transferrin is taken into the cell via endocytosis and localization of labeled transferrin can be imaged in the various intracellular compartments. If a compound inhibits endocytosis, the labeled transferrin will not enter the cell leading to a decrease of fluorescent signal. Inhibitors of interest should act selectively at SR-BI and should have no activity in this assay.
Cells were pre-treated with compound for 3 hours and then treated with the Alexa-594 transferrin reagent for 30 minutes (Catalog No.T-13343; Invitrogen) in serum-free media. Plates of cells were then placed onto ice, washed with ice cold PBS, and then fixed with 4% paraformaldehyde. In addition, the nuclei were stained with 300 nM 4′,6-diamidino-2-phenylindole (DAPI) (Catalog No. 21490; Invitrogen). The cells were imaged with the Molecular Devices IXM microscope. Translocation measurements were performed using MetaExpress software computations and normalized for cell number.
2.1.9. DiI-HDL Uptake Assay with Washout (AID 588831)
This is a modification of the primary assay where cells are pretreated with compound but no compound is present during the DiI-HDL treatment period. ldlA[mSR-BI] cells were plated into 384-well plates at 30 µl per well and incubated overnight. As a measurable surrogate for cholesterol uptake, human HDL particles were treated with the lipophilic fluorescent dye DiI and exposed to ldlA[mSR-BI] cells in lipoprotein-free media (Ham's F12/0.5% fatty acid-free Bovine Serum Albumin [BSA]/25 mM HEPES pH 7.4 plus 10 µg protein/ml DiI-HDL). Cells were treated with compound for 3 hours prior to addition of DiI-HDL. Media were removed, cells were washed with PBS and fresh lipoprotein-free media was added with 10 µg protein/ml DiI-HDL. Cells took up the Dil via SR-BI over 3 hours in the absence of compound. After significant uptake of the DiI, the cells became fluorescent. The level of fluorescence correlates with the amount of Dil uptake and can be measured with a standard plate reader. The uptake of lipid (represented by DiI) was inhibited by the compound BLT-1.
The ldlA[mSR-BI] cells used in the assay were a CHO cell line lacking expression of the LDL receptors and overexpressing the Scavenger receptor (SR-BI). Inhibitors of SR-BI and HDL-mediated uptake lead to a reduction in fluorescence in this assay. Fluorescence was measured using a PerkinElmer EnVision plate reader. Primary HTS data was analyzed in Genedata Assay Analyzer, and were normalized against DMSO and 1 µM BLT-1 (positive control). For the HTS, the average of two replicates was used to rank order activity and to choose compounds for retests. For dose studies, the curves were generated with Genedata Condeseo and showed percent (%) activity for the individual doses.
2.1.10. DiI-HDL Uptake Assay with Extended Washout (AID 588831)
This is another modification of the primary assay where cells are pretreated but no compound is present during a 4-hour washout period and the 3-hour DiI-HDL treatment period. ldlA[mSR-BI] cells were plated into 384-well plates at 30 µl per well and incubated overnight. As a measurable surrogate for cholesterol uptake, human HDL particles were treated with the lipophilic fluorescent dye DiI and exposed to ldlA[mSR-BI] cells in lipoprotein-free media (Ham's F12/0.5% fatty acid-free Bovine Serum Albumin [BSA]/25 mM HEPES pH 7.4 plus 10 µg/ml DiI-HDL). Cells were treated with compound for 3 hours prior to addition of DiI-HDL. Cells were washed four times with PBS, washed once with lipoprotein-free media and returned to the incubator with lipoprotein-free media for 4 hours. Cells took up the Dil via SR-BI over an additional 3 hours in the absence of compound for a total of 7 hours. After significant uptake of the DiI, the cells became fluorescent. The level of fluorescence correlates with the amount of Dil uptake and can be measured with a standard plate reader. The uptake of lipid (represented by DiI) was inhibited by the compound BLT-1 or with an excess of HDL untreated with DiI.
The ldlA[mSR-BI] cells used in the assay were a CHO cell line lacking expression of the LDL receptors and overexpressing the Scavenger receptor (SR-BI). Inhibitors of SR-BI and HDL-mediated uptake lead to a reduction in fluorescence in this assay. Fluorescence was measured using a PerkinElmer EnVision plate reader. Primary HTS data was analyzed in Genedata Assay Analyzer, and were normalized against DMSO and 1 µM BLT-1 (positive control). For the HTS, the average of two replicates was used to rank order activity and to choose compounds for retests. For dose studies, the curves were generated with Genedata Condeseo and showed percent (%) activity for the individual doses.
2.1.11. Lipid Transport Assay in Liposomes (AID 602155)
This assay provides extra confirmation of activity via SR-BI by measuring the uptake of [3H]CE into liposomes loaded with purified SR-BI. For the purification of C-terminally epitope-tagged murine SR-BI (mSR-BI-t1) with uniform, truncated N-linked oligosaccharide chains, we overexpressed mSR-BI-t1 in HEK293S cells. The mSR-BI-t1 was expressed in an N-acetylglucosaminyltransferase I (GnTI)-defective HEK293S derivative, HEK293S GnTI (2), which generates a glycoprotein with uniform, truncated N-linked oligosaccharide chains under the control of a tetracycline-inducible promoter. It was immunoaffinity purified to virtual homogeneity, and the detergent-solubilized receptor was reconstituted into liposomes. Subsequently, the in vitro SR-BI-mediated selective uptake of [3H]CE from [3H]CE-HDL in the reconstituted liposomes was measured. The mSR-BI-t1 with truncated N-linked chains was purified and reconstituted into liposomes as described previously (4). Briefly, 20 µg of SR-BI (or an equivalent volume of protein-free buffer to generate control liposomes that are devoid of SR-BI) was reconstituted into liposomes by acetone precipitation. SR-BI liposomes were washed once by resuspension of the acetone precipitate in protein-free assay medium followed by a centrifugation step for 25 min and 48,000 g at 4 ºC. The pellet was first reconstituted in assay medium without protein, and then an equal volume of assay medium with 1% fatty acid -free BSA was added to yield liposomes at a nominal final concentration of 18 ng SR-BI/ml. In each reaction, 30 ml were pre-incubated together with 30 ml of assay medium containing 0.5% fatty Acid-free BSA, 1% DMSO, and the indicated compounds for 60 min at 37 ºC. Subsequently, 20 µl of [3H]CE-HDL (five replicates per sample) were added to a final concentration of 10 µg protein/ml. Incubation was continued for 4 hours at 37 ºC, and then selective uptake of [3H]CE into liposomes was determined using the previously described filter binding assays (4). SR-BI-selective uptake values were determined. The 100% of control value represents receptor-specific activity in SR-BI-t1-containing liposomes in the presence of 0.5% DMSO without compounds, and the 0% of control value represents background selective uptake in control liposomes devoid of SR-BI-t1.
2.2. Probe Chemical Characterization
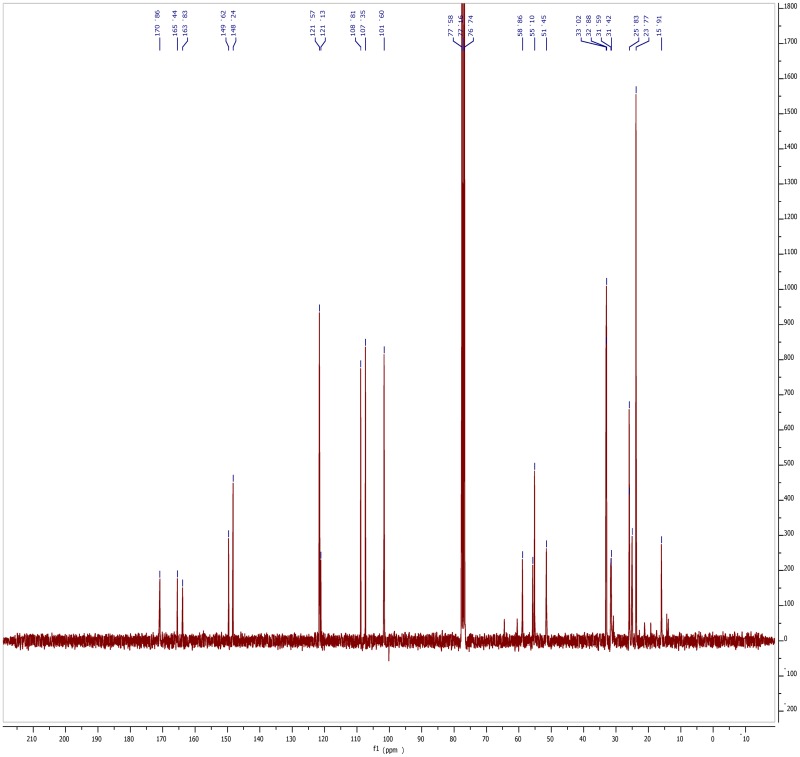
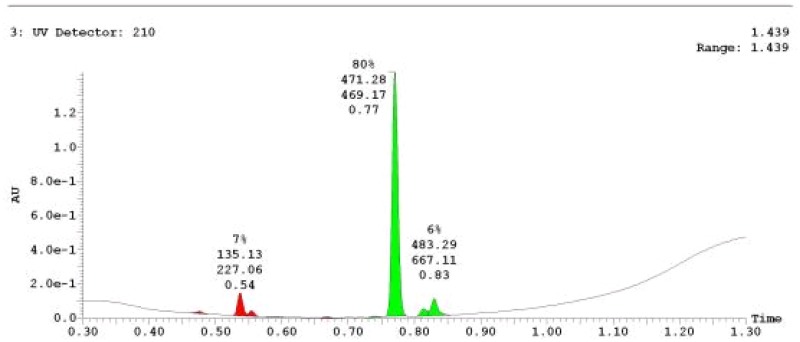
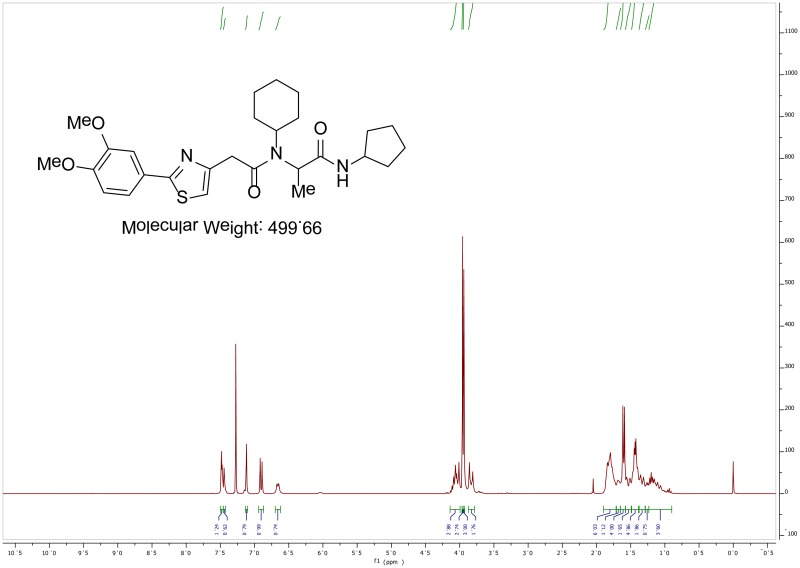
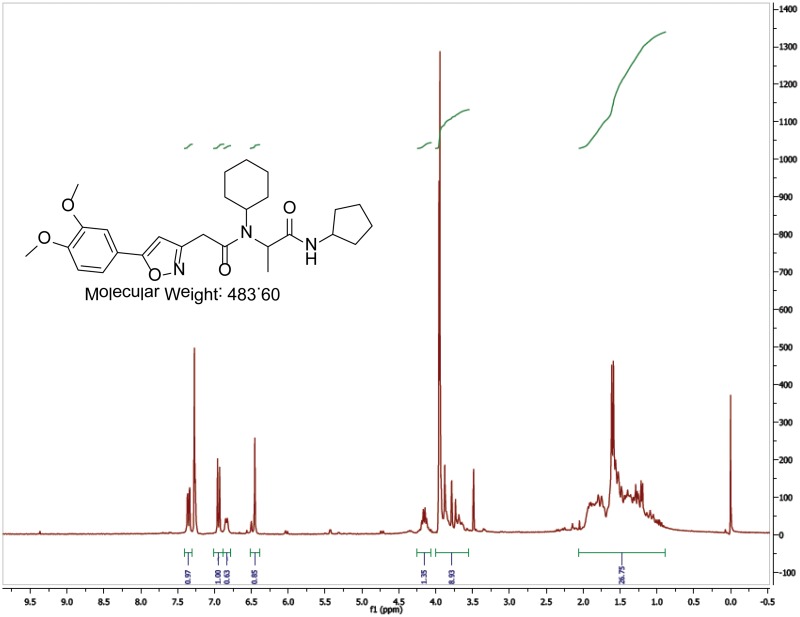
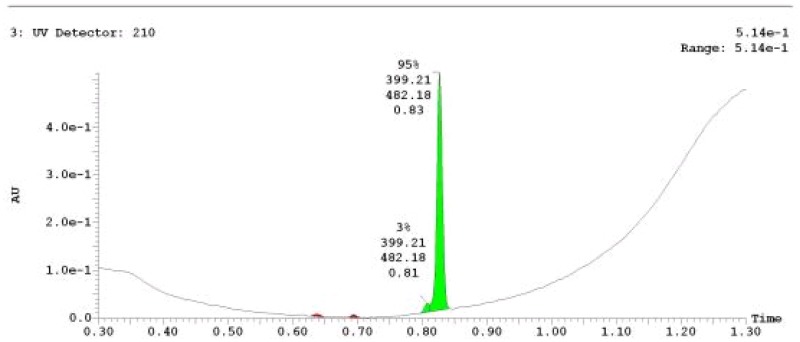
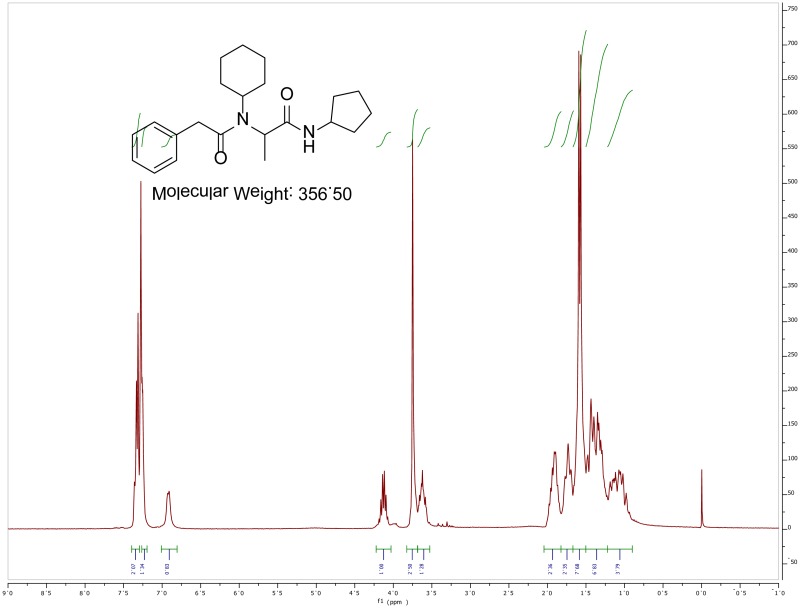
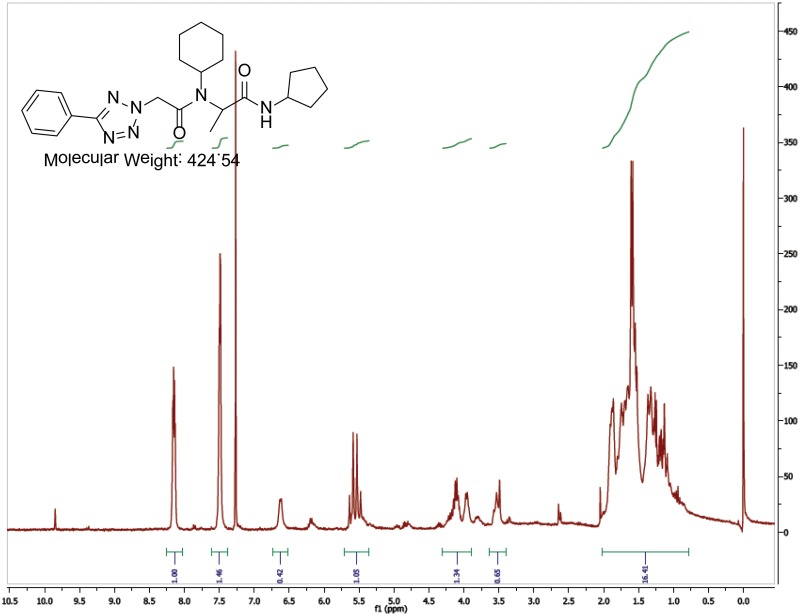
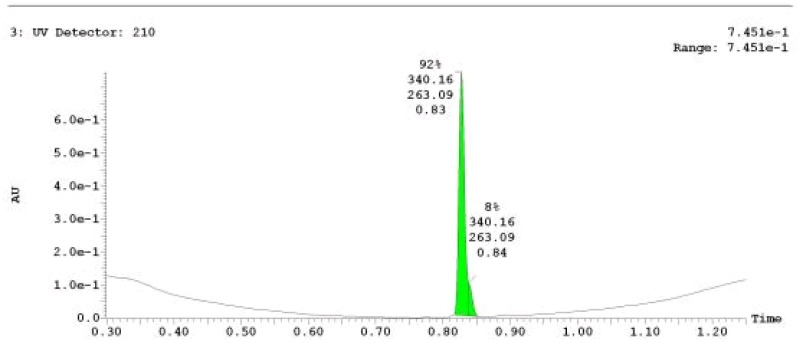
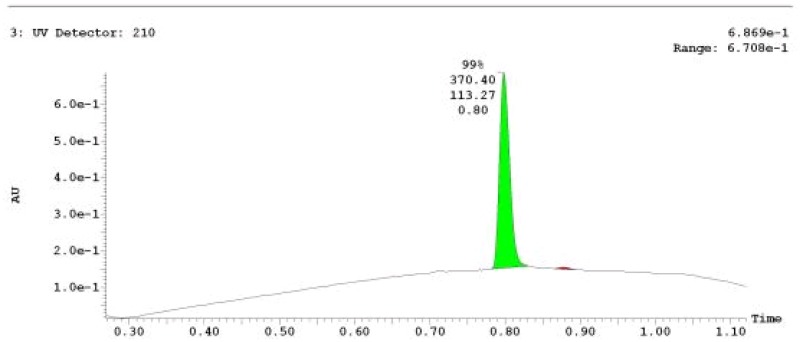
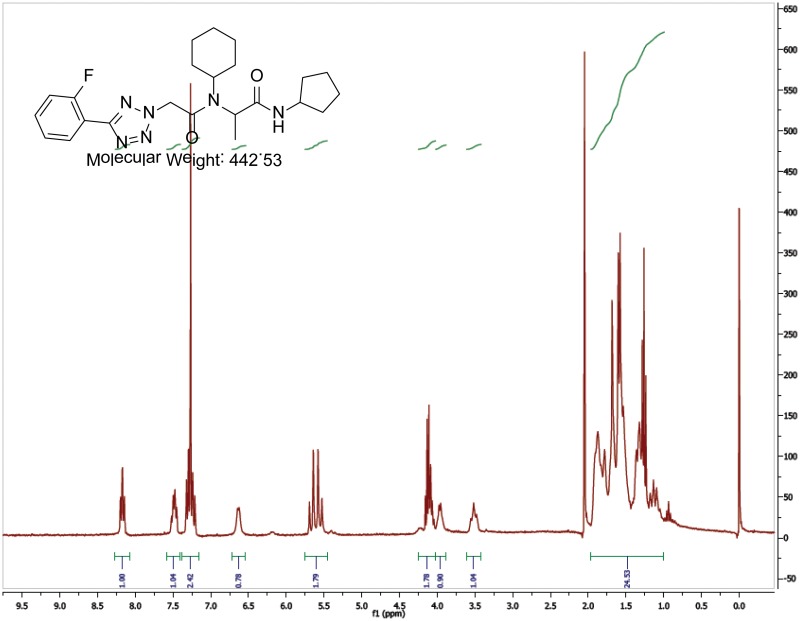
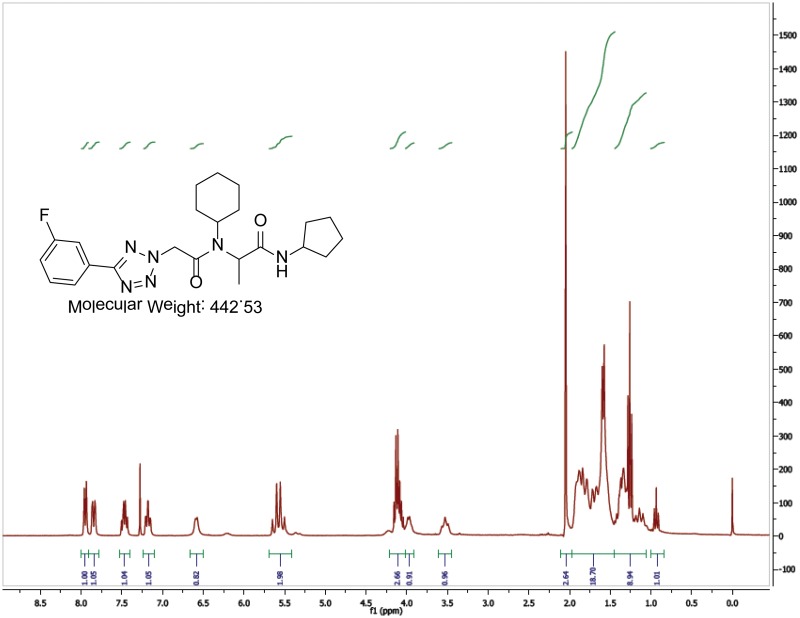
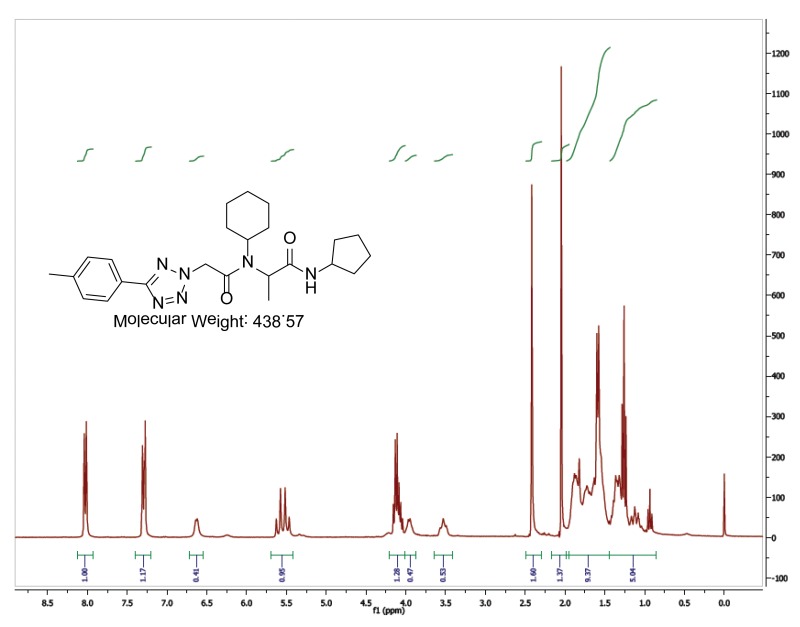
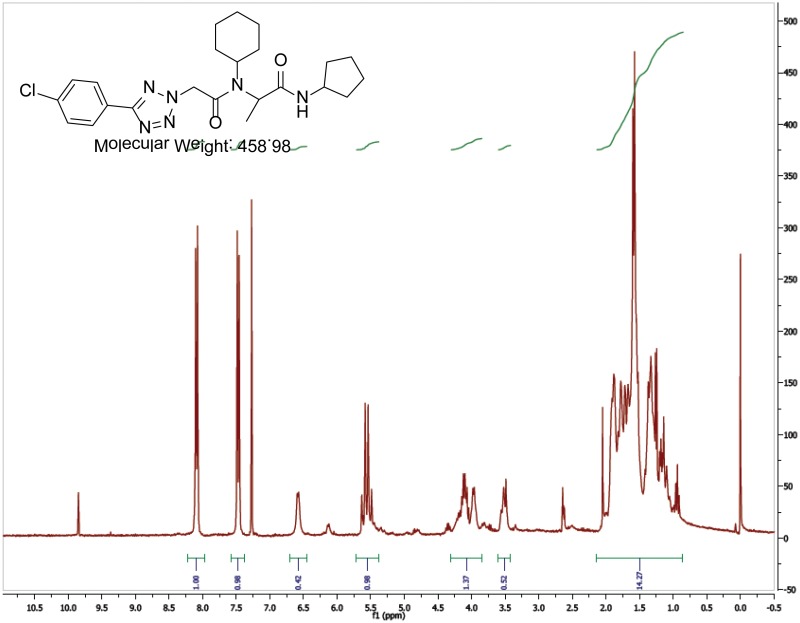
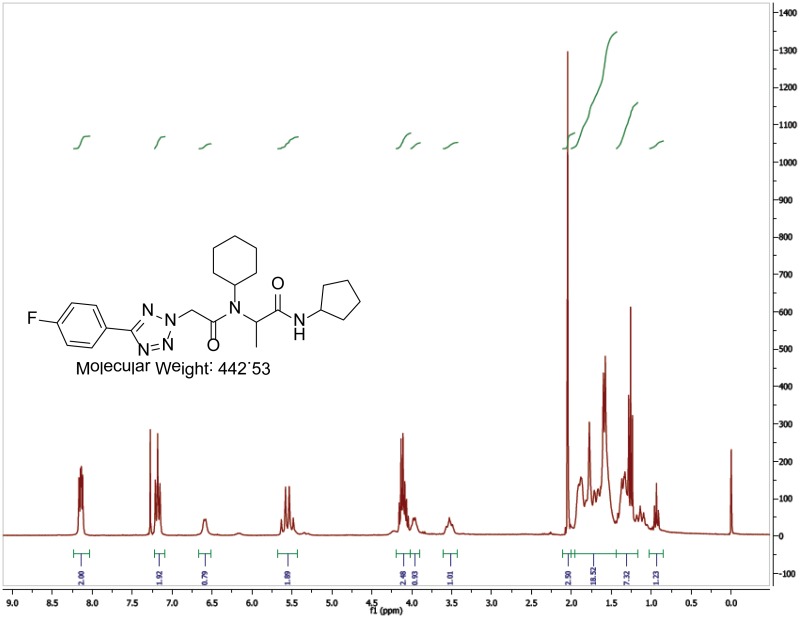
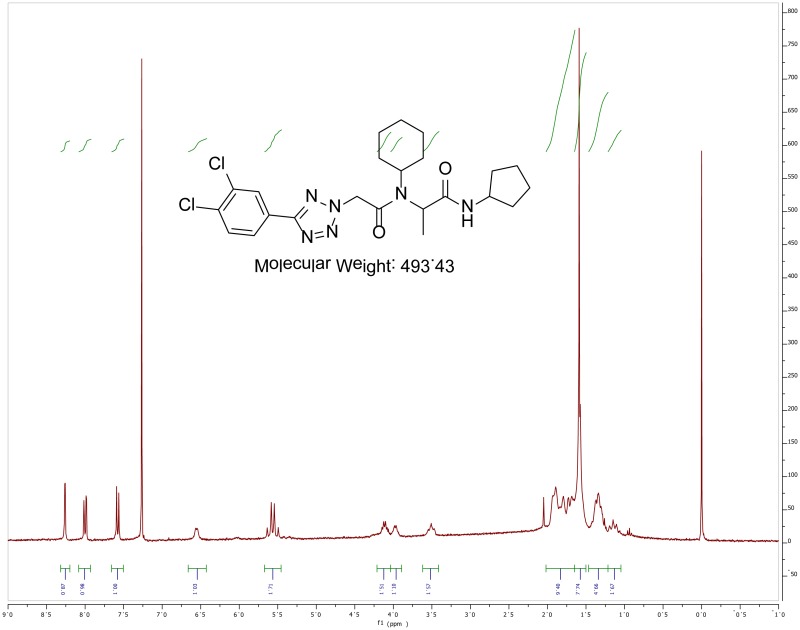
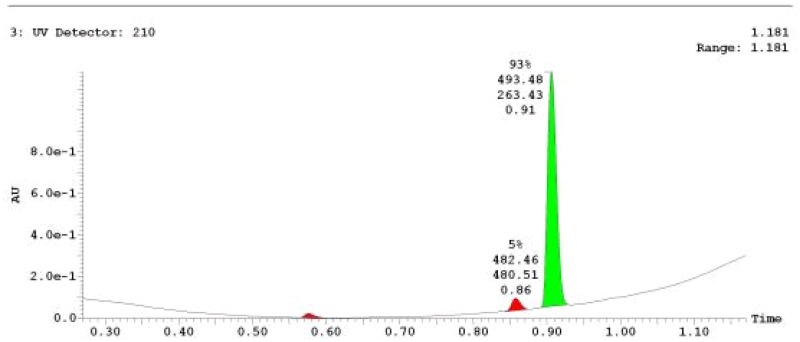
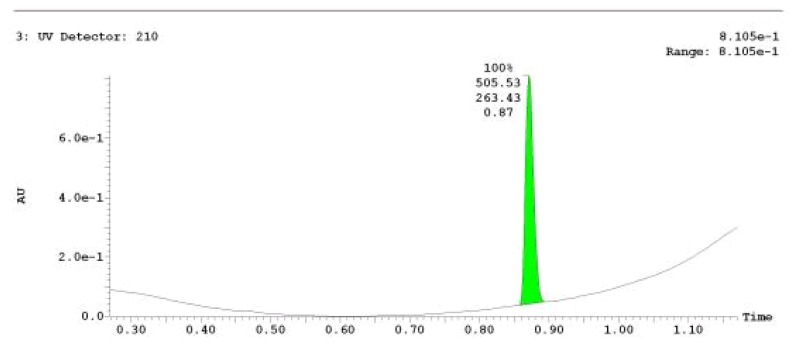
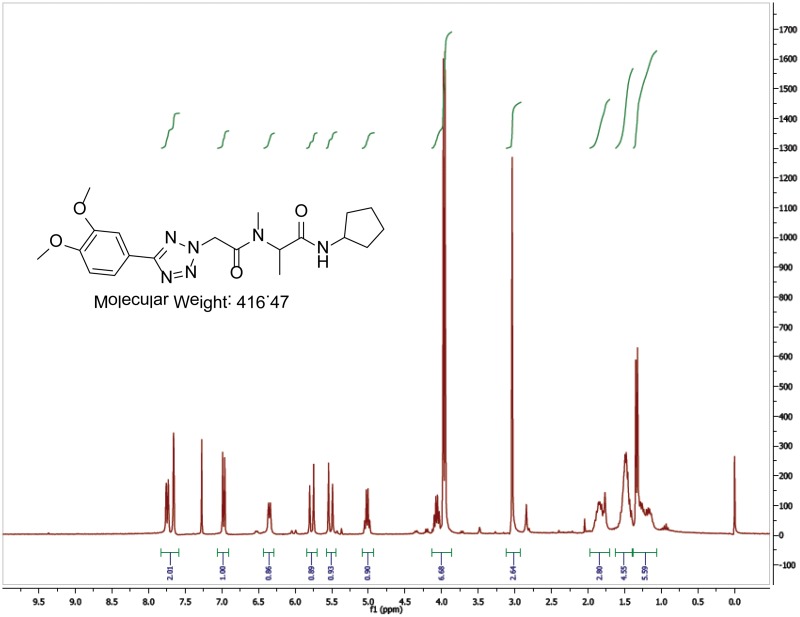
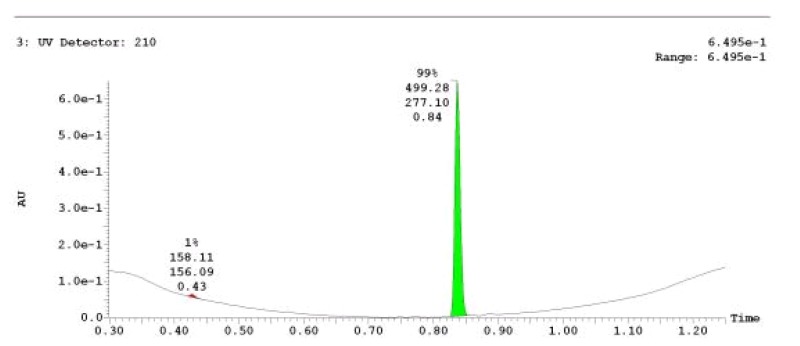
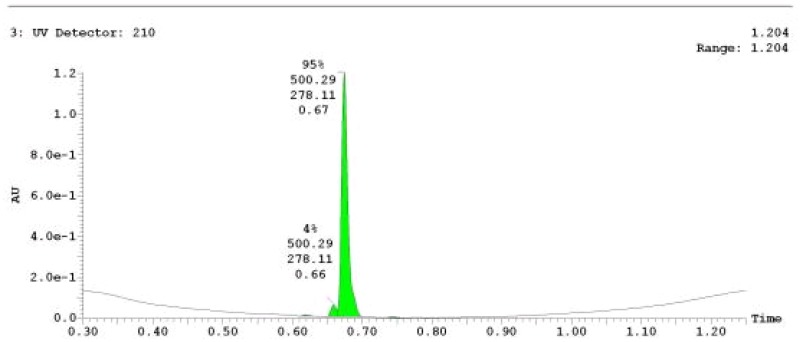
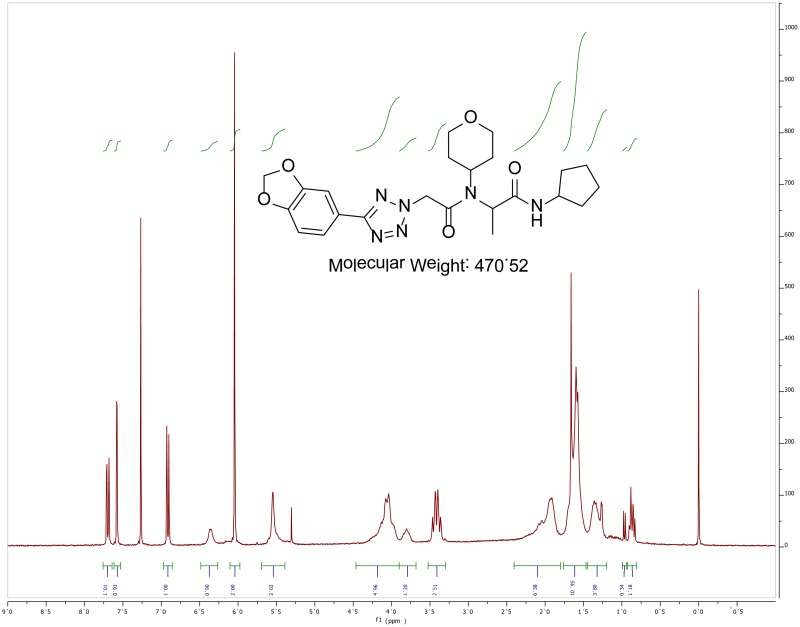
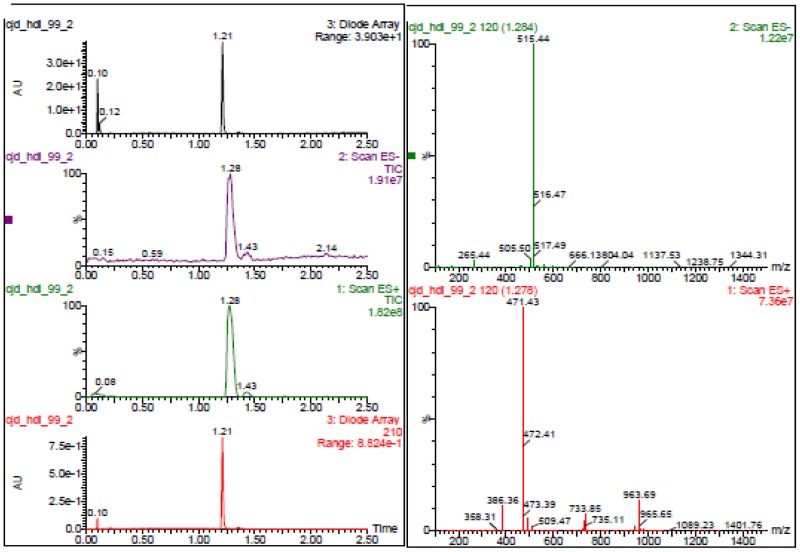
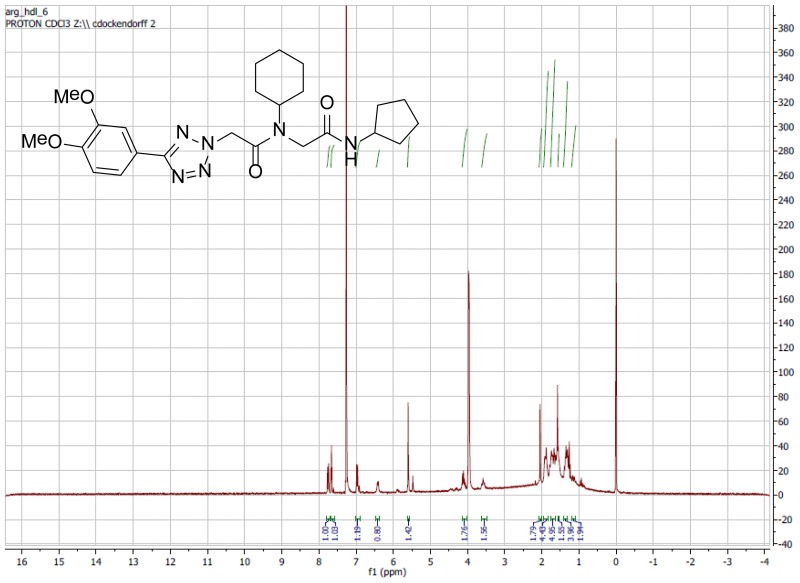
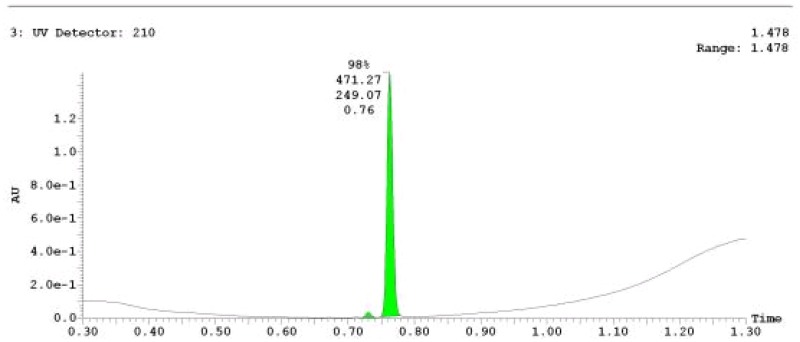
The probe (ML279) was prepared as described in Section 2.3, and was analyzed by UPLC, 1H and 13C NMR spectroscopy, and high-resolution mass spectrometry. The data obtained from NMR and mass spectrometry were consistent with the structure of the probe, and UPLC analysis showed purity of >95%. Characterization data (1H NMR spectra and UPLC chromatograms) of the probe are provided in Appendix E.
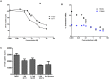
The solubility of Probe 2 (ML279) was determined to be 28 µM in Phosphate buffered saline (PBS; pH 7.4, 23 °C) solution with 1% DMSO. The stability of the probe in PBS (1% DMSO) was measured over 48 hours. We noticed that the concentration of the probe fluctuated through the course of the assay (data not shown). We believe that this fluctuation is due to the low kinetic solubility of the probe. Thus, we decided to determine the amount of undissolved probe present in the well after it was treated with PBS for a given length of time, relative to the amount of undissolved probe at the start of the assay. The wells were centrifuged and the supernatant was removed after various time points, then acetonitrile was added to dissolve the remaining solid in the wells, and the amount of probe was quantified. From these results, the probe seems to be stable to PBS (Figure 2). The stability of the probe was confirmed by measuring stability in human plasma, with >99% remaining after a 5-hour incubation period. Plasma protein binding (PPB) studies showed that it was 76.5% bound in human plasma.
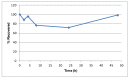
Figure 2
Recovery of Undissolved Probe (ML279) from PBS Buffer (pH 7.4, 23 °C). Percent (%) Recovered is relative to the undissolved solid recovered at T = 0 (100%), quantified by LC-MS.
Experimental procedures for the various analytical assays are provided in Appendix D. Table 1 summarizes known properties of the probe.
Table 1
Summary of Known Probe (ML279) Properties Computed from Structure.
2.3. Probe Preparation
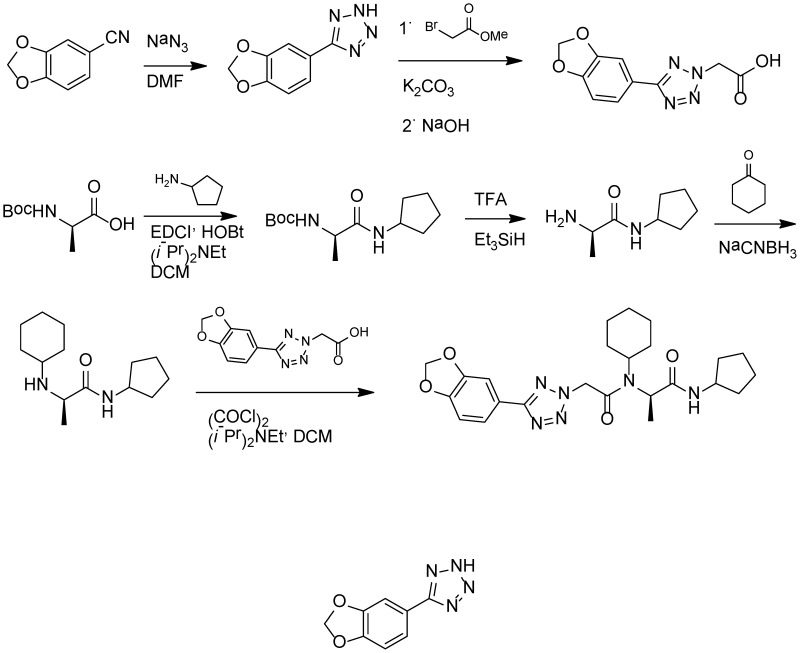
The racemic version of Probe 2 (ML279), and most of the analogs prepared for SAR analysis were conveniently prepared via Ugi 4-component reaction. The enantiopure Probe 2 (ML279) was made using standard peptide synthesis protocols, as shown in Scheme 1. The tetrazole acetic acid building block was prepared by condensing the requisite arylnitrile with sodium azide, then alkylating the resulting tetrazole with methyl bromoacetate, and finally hydrolyzing the ester (Scheme 1). Full experimental details are provided in Appendix C.
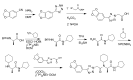
Scheme 1
Synthesis of Probe 2 (ML279).
3. Results
- Has an IC50 value as low as 16 nM in the DiI-HDL uptake assay.
- Shows no apparent cytotoxicity after 24 hours treatment (IC50= >35 µM).
- Inhibits efflux of free cholesterol.
- Does not inhibit endocytosis of transferring.
- Is a reversible inhibitor.
To identify novel inhibitors, SR-BI-mediated lipid uptake was measured in a customized cell line that minimized potential uptake by other mechanisms, including via the LDL receptor (1). This primary cell-based assay was used in a pilot screen that identified BLTs 1 through 5 (2). The assay was slightly modified from the original protocol of Nieland et al. to increase throughput and to ease automation procedures.
Briefly, the plate reader was switched to a Perkin-Elmer EnVision, several washing steps were eliminated, and the cell number per well was reduced without impacting the robustness of the assay. The automated assay was first tested with a validation set of 2,240 compounds. These compounds were tested in duplicate with in-plate neutral (DMSO) and positive controls. Robustness, reproducibility, and variability parameters were analyzed before initiating the full HTS. The HTS was run over the course of several weeks. Data were normalized relative to controls, and plate patterns were corrected using a multiplicative algorithm in the Genedata Assay Analyzer. For each compound, the average of the two replicates was determined and used for subsequent analysis.
Determination of hits required several criteria: only assay plates with a Z′ greater than 0.3 were accepted for analysis, compounds needed to reach 70% inhibition relative to 1 µM BLT-1, and score in fewer than 10% of HTS assays listed in PubChem. In total, 318,714 compounds were screened. Of these, 3,046 compounds were considered active (a hit rate of 0.96%), 613 were inconclusive, and 315,055 were inactive. Compounds were clustered based upon chemical structure using a customized script in Pipeline Pilot. Clusters were rated based upon structural liabilities and ranked accordingly. Substructures were analyzed and compared to inactive compounds to identify inactive analogs. Representatives from the more desirable clusters were selected for retest. In addition, a small number of inactive analogs were chosen to provide initial structure activity relationship (SAR) data during the dose retest studies. Of these, 573 compounds were retested over a range of concentrations to validate activity, and 186 compounds showed dose-dependent inhibition of DiI-HDL uptake.
Since active compounds produce a decrease in signal in the primary assay, confirmation was required that the reduction in fluorescence was not a result of quenching by the compound. Compounds were tested in the presence of DiI-HDL in a cell-free version of the primary assay. Of compounds that were active in the primary assay, 127 active compounds were found to quench the DiI signal and 59 active compounds showed no alteration of fluorescence in the quenching assay.
All available dry powder samples of the remaining, non-quenching compounds were procured from commercial sources and purities were determined. The compounds obtained were then retested in the primary assay and in cell cytotoxicity assays. We wanted to rule out any compounds that were cytotoxic, especially in the 3-hour time span of the primary DiI-HDL uptake assay. Therefore, compounds were tested in [ldl]mSR-BI cells for cytotoxicity at 3 hours and 24 hours using the CellTiter-Glo assay (Promega) that measures cellular ATP as a surrogate marker of cell viability. Of these, 11 compounds showed an IC50 value of less than 35 µM and were excluded from further consideration. Approximately 44 compounds had an IC50 value below 10 µM in the DiI-HDL assay. Fifteen chemical scaffolds showed potencies below 1 µM and three of these were prioritized for follow-up studies. Compounds were screened for inhibiting endocytosis using labeled transferrin, which is known to bind the transferrin receptor, enter cells by clathrin-mediated endocytosis, and move to endosomes before being recycled to the cell surface. The BLT compounds have been tested for potential interference with endocytosis and show no such activity (2).
The original tetrazole-peptide hit described in this report, the peptide scaffold MLS000888573 (Figure 3, SID 26667991, CID 650573), did not alter endocytosis of transferrin (Figure 10B, AID 602124). With an IC50 value in the primary assay of 0.051 µM (Figure 4, AID 540354), the peptide scaffold of MLS000888573 was thus prioritized for further development. Analogs were designed and synthesized, eventually leading to the more potent probe, ML279 (see Section 3.4).
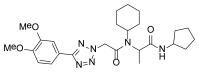
Figure 3
The Tetrazole Peptide MLS000888573.
3.1. Summary of Screening Results
Refer to subsections for a detailed description of the results.
In the primary HTS, compounds were active if they decreased DiI-HDL uptake as measured by fluorescence. The positive control, BLT-1 (1 µM), caused a decrease in uptake that led to over a 2.2-fold reduction in signal. Compounds with greater than 70% activity of the positive control were considered actives and chosen for confirmation studies (see Section 3.4 for details).
Figure 5 displays the critical path for probe development. To explore SAR, a number of analogs were synthesized and tested. Selected results are shown in Tables 2-7 in Section 3.4.
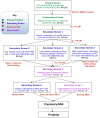
Figure 5
Critical Path for Probe Development.
Table 2
SAR Analysis of Eastern Amide.
Table 7
SAR Analysis of Central Carbon Substituent.
3.2. Dose Response Curves for Probes
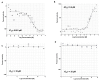
Figure 6ML279 was used over a range of concentrations up to 35 μM in the primary assay
DiI-HDL uptake assay IC50 = 0.0162 µM (AID 588828) (A); Alexa-488 HDL binding assay, EC50 = 0.27 µM (AID 588810) (B); DiI-HDL ldlA7 counterscreen, IC50 >35 µM (AID 588825) (C); 24-h CellTiter-Glo cytotoxicity, IC50 >35 µM (AID 588829) (D). Dose curves were generated with Genedata Condeseo and show normalized percent activity for the individual doses. ○=replicate 1, △=replicate 2
3.3. Scaffold/Moiety Chemical Liabilities
As discussed in Section 2.2, Probe 2 (ML279) could potentially act as an electrophile under biological conditions, with its tetrazole alpha to the western amide. Currently, there is no evidence to support this possibility, as it is stable to GSH, and the racemic version of the probe showed reversible inhibition of SR-BI mediated lipid uptake. Modified versions of the DiI-HDL protocol were used to allow ML279 to wash out for either the 3-hour duration when the cells are taking up the DiI-HDL, or for a 4-hour period prior to the DiI-HDL addition. In both cases, the observed activity was greatly attenuated, indicating that ML279 was being washed away and was not reacting with the cells in an irreversible manner (AID Nos. 588831, 602154).
3.4. SAR Tables
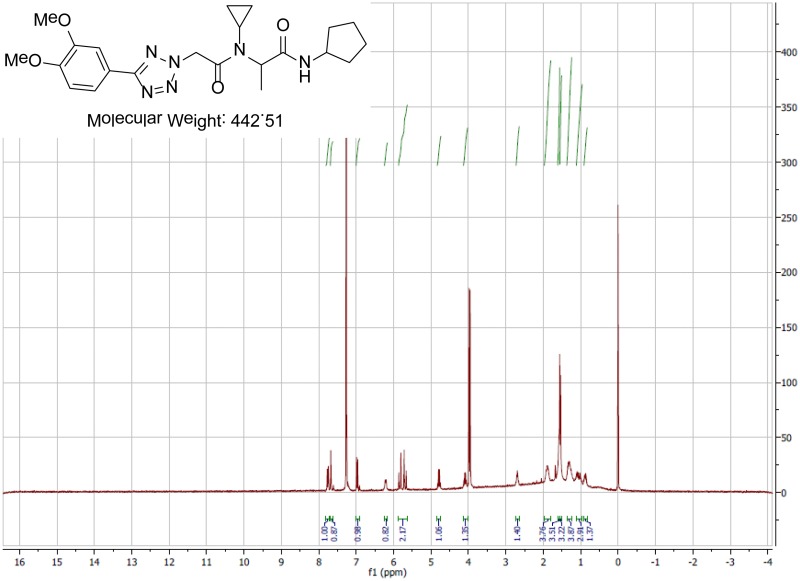
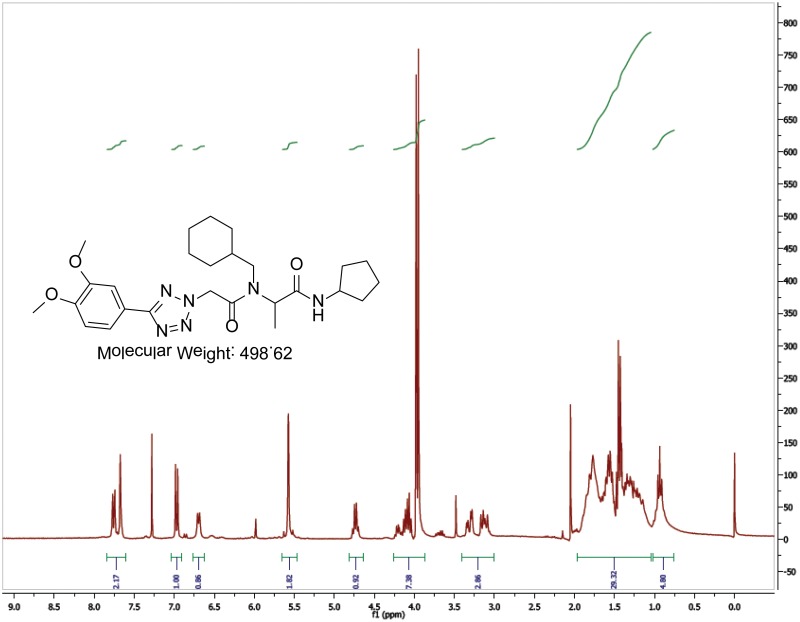
The biological assay data and physical properties of these analogs are presented in Tables 2-7. Characterization data (1H NMR spectra and UPLC chromatograms) of these analogs are provided in Appendix H. Data on additional analogs is included in Appendix I.
The hit compound (Tables 2 to 6, Entry 1) showed good potency (IC50 = 0.058 μM) and excellent solubility in PBS (114 μM). In order to rapidly explore the SARs at different parts of the molecule, we generally utilized the 4-component Ugi reaction, which could often provide the desired products in a single reaction from appropriate building blocks (aldehyde, amine, carboxylic acid, and isonitrile). We investigated the eastern amide portion of the molecule by varying the isonitrile building block. The SAR at this location is very steep, as smaller (Table 2, Entries 2, 3) or larger (Table 2, Entry 4) terminal alkyl groups largely abrogated the activity. A secondary amide is required for optimal activity, as structurally similar tertiary amides had greatly reduced potencies (Table 2, Entries 5, 6). Both tertiary amides were prepared via a stepwise peptide synthesis; N-alkylation of the hit compound gave a complex mixture due to competing C-alkylation alpha to the tetrazole.
Table 6
SAR Analysis of Probe 2 Central Amide: N-substituent.
Several replacements of the tetrazole ring were investigated (Table 3), using arylacetic acid building blocks in the Ugi reaction that were purchased or prepared in several steps. An oxazole (Table 3, Entry 2) showed decent potency, roughly two-fold less that the hit compound (IC50 = 0.13 μM). A thiazole replacement (Table 3, Entry 3) had poor activity, but an isoxazole (Table 3, Entry 4) had excellent potency in the primary assay (IC50 = 0.023 μM) and good solubility. A highly simplified arene analog without a heterocycle (Table 3, Entry 5) was basically inactive.
Table 3
SAR Analysis of Heterocycle: Tetrazole Replacements.
The arene SAR at the western end of the scaffold was investigated next (Tables 4, 5). Several ortho-substituted arenes were poorly active (Table 4, Entries 4, 5). Two electron-donating groups (or possibly H-bond acceptors) proved to be optimal, as removal of the 4-methoxy group (Table 4, Entry 3) decreased the activity approximately 3-fold. Pleasingly, replacement of the 3,4-dimethoxy groups with a methylenedioxy acetal (Table 5, Entry 6) gave a slight increase in activity (IC50 = 0.037 μM), and we expect will likely also give improved metabolic stability (results pending). The difluoromethylenedioxy analog was prepared but possessed significantly less activity (Table 5, Entry 7).
Table 4
SAR Analysis of Probe 2 Western Arene: Ortho and Meta Substitutions.
Table 5
SAR Analysis of Probe 2 Western Arene: Para Substitutions.
Variation of the primary amine utilized in the Ugi reaction provided access to compounds with various nitrogen substituents at the central amide (Table 6). The SAR again proved to be fairly steep, as smaller groups such as methyl and cyclopropyl (Table 6, Entries 2-4) had decreased activity. Introduction of a methylene spacer between the cyclohexane group and the nitrogen (Table 6, Entry 5) largely abolished the activity. Introduction of heteroatoms to the cyclohexane ring (Table 6, Entries 6, 7) also decreased the activity significantly. The Ugi reaction using aniline as the amine component (to provide the desired N-phenyl substituent) failed using standard conditions, though we expect that it could be prepared if desired using alternative conditions.
Finally, we investigated substitution at the central carbon of the peptide (Table 7). Surprisingly, addition of an additional methyl group abolished the activity (Table 7, Entry 1), as did replacement of the methyl group with a bulkier isopropyl group (Table 7, Entry 2). The use of formaldehyde in the Ugi reaction provided achiral and unsubstituted analogs (Table 7, Entry 4), which can be compared to the prior top compound (Table 7, Entry 3). Both examples show similar potency to the best analogs, though the solubility is significantly lower (as low as 3.7 μM in PBS; Table 7, Entry 4). To investigate the importance of the chiral center in the methyl-substituted analogs, the individual enantiomers of our racemic lead compound (Table 7, Entry 3) were prepared from the D- and L-isomers of alanine (see Section 2.3). The chirality of the molecule is indeed important, as the bulk of the activity resides in the (R)-isomer (Table 7, Entry 6, IC50 = 0.018 μM, versus IC50 = 1.6 μM for the (S)-isomer (Table 7, Entry 5). With optimal potency and decent solubility (27 μM in PBS), we, thus, nominated CID 53393835 as the probe for this series of HDL-mediated lipid uptake inhibitors.
3.5. Cellular Activity
The primary assay and several of the secondary assays are cell-based experiments. The compound shows activity in cells and the effective IC50 value for the probe in the primary assay (0.018 µM) is well below the measured PBS solubility (28 µM). This inhibition of uptake was confirmed with an IC50 value of 1 µM in a nonfluorescent, radiolabeled version of the assay (Figure 9A, AID 588836). Since SR-BI is a cell-surface receptor (and the compound is presumed to act on the extracellular surface), cell permeability is not an issue, though the probe is expected to have reasonable permeability. ML279 showed no cytotoxicity at 24 hours (Figure 6C, AID 540326). The precise mode of action for the probe is not known but several supplemental assays suggest a selective and direct interaction with SR-BI (see Mechanism of Action, Section 4.2). The probe shows no effect on the endocytosis of transferrin, at concentrations up to 35 µM (Figure 10B, AID 602134).
3.6. Profiling Assays
The probe will be sent to Ricerca for evaluation of off-target binding to a broad panel of receptors, ion channels and enzymes. Metabolic stability will also be evaluated.
4. Discussion
The HTS campaign described in Section 3.1 of a library of small molecules identified a hit that was modular and amenable to medicinal chemistry studies. We prepared numerous synthetic analogs and investigated structure-activity relationships (SARs; Figure 7) at five different parts of the peptide scaffold, leading to the identification of the probe (ML279).
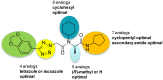
Figure 7
Summary of SAR Performed.
Currently, the probe is not in listed in PubChem but it was modified from the original HTS active compound, which is listed under two records (MLS000077291 and MLS000888573) and three SIDs.
According to PubChem, MLS000077291/MLS000888573 (SID 26667991, SID 848144, SID 115950079, CID 650573) has been tested in 614 bioassays and was active in four assays excluding inhibition of SR-BI uptake (AID 696, AID 777, AID 778, and AID 504444). There was no dose retest reported for these projects as of the date of submission for this probe report except for the Nrf2 qHTS screen for inhibitors (AID 504444). In this assay, the compound showed a partial inhibition that was categorized as “unspecified” results. Therefore, we expect the probe to have good selectivity over other biological targets. This selectivity will be confirmed by a Ricerca off-target binding screen.
In summary, 36 analogs were prepared and screened, leading to a probe with improved potency over the original hit and a reduced metabolic liability (through replacement of the dimethoxybenzene group with a methylenedioxybenzene).
Table 8 presents a comparison of the activity and selectivity profile of the probe (ML279) with the probe criteria decided upon in the chemical probe development plan (CPDP).
Table 8
Comparison of the Probe to Project Criteria.
4.1. Comparison to existing art and how the new probe is an improvement
Several prior art compounds were compared to ML279 and the properties of the probe exceed the prior art (Figure 8, AID 588828). BLT-1 is a potent inhibitor of SR-BI-mediated lipid uptake and of free cholesterol efflux. It is a nonreversible covalent modifier of SR-BI and is toxic to cells. ML279 does not have any structural motifs that suggest covalent modification; it is a reversible inhibitor of HDL uptake (AID 588831, AID 602154) and shows no cytotoxicity in [ldlA] mSR-BI cells (Figure 6C, AID 588829). Another inhibitor of SR-BI is ITX-5061. It was originally identified as a p38 MAPK inhibitor that was found to elevate circulating levels of HDL-cholesterol in human patients (6). We synthesized ITX-5061 and tested it in multiple assays. In every experiment, ML279 outperformed ITX-5061, with approximately 15-fold higher potency (Figure 8, AID 588828). As discussed in Section 1, ITX-5061 is also a known MAP kinase inhibitor, and it contains a ketoamide functionality that may be susceptible to covalent reaction with various targets.
Investigation into relevant prior art entailed searching the following databases: SciFinder, Web of Science, PubMed, and Patent Lens. The search terms applied and hit statistics for the prior art search are provided in Table A2 (Appendix G). Abstracts were obtained and were analyzed for relevance to the current project. The searches are current as of November 26, 2011.
Table A2
Search Strings and Databases Employed in the Prior Art Search.
4.2. Mechanism of Action Studies
ML279 and selected analogs were tested in several secondary assays that address possible mechanisms of action. In addition to uptake of HDL particles and bringing esterified cholesterol into the cell, SR-BI also plays a role in the efflux of free cholesterol from the cell to recipient HDL particles. Compounds were tested to determine if there was an impact on efflux from cells. Similar to BLT-1, ML279 reduces efflux of free cholesterol from cells by about 50% (Figure 9B, AID 602152). Unlike BLT-1, ML279 was shown to be a reversible inhibitor in the DiI uptake assay, In experiments where cells were pre-treated with compounds for 2 hours, washed extensively with PBS and then incubated with DiI-HDL without compound, ML279 showed no inhibition (AID 588831). In comparison, BLT-1 registers an IC50 value of 600 pM even after a 4-hour washout period (AID 602154), reflecting the progression of its covalent reaction with cysteine 384 of SR-BI (5). ML279 was tested for efflux in a cell line that only expresses a mutant form of the protein where Cys384 is converted to a serine residue. In this mutant background, BLT-1 cannot reduce uptake of HDL or efflux of cholesterol. ML279 can reduce uptake and efflux in the mutant background suggesting that the compound works at a different site of the receptor and does not involve Cys384. We assessed binding of HDL to SR-BI using an Alexa-488 labeled HDL. Similar to BLT-1, ML279 increases binding of HDL to the receptor (Figure 6D, AID 588810). Further studies need to be performed to determine if this is a result of tighter binding or inhibition of the disassociation of HDL (2).
Outside the scope of the CPDP, MLS000888573 was tested with SR-BI in purified liposomes. These data suggest a direct interaction between the probe and SR-BI (Figure 9C, AID 602155). This data substantiates results in [ldlA7] cells where SR-BI is required for compound activity (Figure 6B, AID 588825).
4.3. Planned Future Studies
Extensive medicinal chemistry was performed to generate ML279, a highly potent SR-BI inhibitor. Some additional medicinal chemistry work is in progress to produce new analogs with improved solubility, though deadlines dictated that this work could not be described here. We will also examine the surface expression levels of the SR-BI protein. Much work remains to be done to elucidate the specific interactions between the probe and SR-BI. These studies will be performed by the Assay Provider. The liposome studies demonstrate that there is direct interaction between the receptor and ML279, but the specific site of interaction remains to be determined. Multiple mutant versions of the receptor are available and will help to identify what portion of SR-BI is needed for interaction. SR-BI has been shown to dimerize, and ML279 will be tested with various dimerization-incompetent forms of the receptor to determine if the compounds interfere with dimerization (19). In addition, future studies could investigate if the probe prevents interaction with other proteins that are known to interact with SR-BI (20). These studies depend upon knowledge of the site of interaction on SR-BI.
Our assays have been focused on SR-BI relative to cholesterol metabolism. SR-BI has roles in immunity and acts as a co-receptor for malaria and hepatitis C virus infection. We plan to test ML279 for its ability to reduce or prevent infection of cells with these two pathogens. Compounds will be tested in collaboration with researchers at the National Institute of Allergy and Infectious Diseases (NIIAID) and at the Broad Institute.
5. References
- 1.
- Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 1996 Jan 26;271(5248):518–20. [PubMed: 8560269]
- 2.
- Nieland TJ, Penman M, Dori L, Krieger M, Kirchhausen T. Discovery of chemical inhibitors of the selective transfer of lipids mediated by the HDL receptor SR-BI. Proc Natl Acad Sci U S A. 2002 Nov 26;99(24):15422–7. Epub 2002 Nov 18. [PMC free article: PMC137732] [PubMed: 12438696]
- 3.
- Nieland TJ, Chroni A, Fitzgerald ML, Maliga Z, Zannis VI, Kirchhausen T, Krieger M. Cross-inhibition of SR-BI- and ABCA1-mediated cholesterol transport by the small molecules BLT-4 and glyburide. J Lipid Res. 2004 Jul;45(7):1256–65. Epub 2004 Apr 21. [PubMed: 15102890]
- 4.
- Nieland TJ, Shaw JT, Jaipuri FA, Maliga Z, Duffner JL, Koehler AN, Krieger M. Influence of HDL-cholesterol-elevating drugs on the in vitro activity of the HDL receptor SR-BI. J Lipid Res. 2007 Aug;48(8):1832–45. Epub 2007 May 28. [PubMed: 17533223]
- 5.
- Yu M, Romer KA, Nieland TJ, Xu S, Saenz-Vash V, Penman M, Yesilaltay A, Carr SA, Krieger M. Exoplasmic cysteine Cys384 of the HDL receptor SR-BI is critical for its sensitivity to a small-molecule inhibitor and normal lipid transport activity. Proc Natl Acad Sci U S A. 2011 Jul 26;108(30):12243–8. Epub 2011 Jul 11. [PMC free article: PMC3145699] [PubMed: 21746906]
- 6.
- Masson D, Koseki M, Ishibashi M, Larson CJ, Miller SG, King BD, Tall AR. Increased HDL cholesterol and apoA-I in humans and mice treated with a novel SR-BI inhibitor. Arterioscler Thromb Vasc Biol. 2009 Dec;29(12):2054–60. Epub 2009 Oct 8. [PMC free article: PMC2783626] [PubMed: 19815817]
- 7.
- Rigotti A, Miettinen HE, Krieger M. The role of the high-density lipoprotein receptor SR-BI in the lipid metabolism of endocrine and other tissues. Endocr Rev. 2003 Jun;24(3):357–87. [PubMed: 12788804]
- 8.
- Fioravanti J, Medina-Echeverz J, Berraondo P. Scavenger receptor type B, class I: A promising immunotherapy target. Immunotherapy. 2011 Mar;3(3):395–406. [PubMed: 21395381]
- 9.
- Guo L, Song Z, Li M, Wu Q, Wang D, Feng H, Bernard P, Daugherty A, Huang B, Li XA. Scavenger receptor BI protects against septic death through its role in modulating inflammatory response. J Biol Chem. 2009 Jul 24;284(30):19826–34. Epub 2009 Jun 2. [PMC free article: PMC2740408] [PubMed: 19491399]
- 10.
- Zhu P, Liu X, Treml LS, Cancro MP, Freedman BD. Mechanism and regulatory function of CpG signaling via scavenger receptor B1 in primary B cells. J Biol Chem. 2009 Aug 21;284(34):22878–87. Epub 2009 Jun 19. [PMC free article: PMC2755695] [PubMed: 19542230]
- 11.
- Catanese MT, Graziani R, von Hahn T, Moreau M, Huby T, Paonessa G, Santini C, Luzzago A, Rice CM, Cortese R, Vitelli A, Nicosia A. High-avidity monoclonal antibodies against the human scavenger class B type I receptor efficiently block hepatitis C virus infection in the presence of high-density lipoprotein. J Virol. 2007 Aug;81(15):8063–71. Epub 2007 May 16. [PMC free article: PMC1951280] [PubMed: 17507483]
- 12.
- Catanese MT, Ansuini H, Graziani R, Huby T, Moreau M, Ball JK, Paonessa G, Rice CM, Cortese R, Vitelli A, Nicosia A. Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants. J Virol. 2010 Jan;84(1):34–43. [PMC free article: PMC2798406] [PubMed: 19828610]
- 13.
- Voisset C, Callens N, Blanchard E, Op De Beeck A, Dubuisson J, Vu-Dac N. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J Biol Chem. 2005 Mar 4;280(9):7793–9. Epub 2005 Jan 4. [PubMed: 15632171]
- 14.
- Syder AJ, Lee H, Zeisel MB, Grove J, Soulier E, Macdonald J, Chow S, Chang J, Baumert TF, McKeating JA, McKelvy J, Wong-Staal F. Small molecule scavenger receptor BI antagonists are potent HCV entry inhibitors. J Hepatol. 2011 Jan;54(1):48–55. Epub 2010 Aug 21. [PubMed: 20932595]
- 15.
- Rodrigues CD, Hannus M, Prudêncio M, Martin C, Gonçalves LA, Portugal S, Epiphanio S, Akinc A, Hadwiger P, Jahn-Hofmann K, Röhl I, van Gemert GJ, Franetich JF, Luty AJ, Sauerwein R, Mazier D, Koteliansky V, Vornlocher HP, Echeverri CJ, Mota MM. Host scavenger receptor SR-BI plays a dual role in the establishment of malaria parasite liver infection. Cell Host Microbe. 2008 Sep 11;4(3):271–82. [PubMed: 18779053]
- 16.
- Yalaoui S, Huby T, Franetich JF, Gego A, Rametti A, Moreau M, Collet X, Siau A, van Gemert GJ, Sauerwein RW, Luty AJ, Vaillant JC, Hannoun L, Chapman J, Mazier D, Froissard P. Scavenger receptor BI boosts hepatocyte permissiveness to Plasmodium infection. Cell Host Microbe. 2008 Sep 11;4(3):283–92. [PubMed: 18779054]
- 17.
- Nieland TJ, Shaw JT, Jaipuri FA, Duffner JL, Koehler AN, Banakos S, Zannis VI, Kirchhausen T, Krieger M. Identification of the molecular target of small molecule inhibitors of HDL receptor SR-BI activity. Biochemistry. 2008 Jan 8;47(1):460–72. Epub 2007 Dec 8. [PMC free article: PMC2736594] [PubMed: 18067275]
- 18.
- Nishizawa T, Kitayama K, Wakabayashi K, Yamada M, Uchiyama M, Abe K, Ubukata N, Inaba T, Oda T, Amemiya Y. A novel compound, R-138329, increases plasma HDL cholesterol via inhibition of scavenger receptor BI- mediated selective lipid uptake. Atherosclerosis. 2007 Oct;194(2):300–8. Epub 2006 Dec 12. [PubMed: 17166497]
- 19.
- Gaidukov L, Nager AR, Xu S, Penman M, Krieger M. Glycine dimerization motif in the N-terminal transmembrane fomain of the high density lipoprotein receptor SR-BI required for normal receptor oligomerization and lipid transport. J Biol Chem. 2011 May 27;286(21):18452–64. Epub 2011 Mar 25. [PMC free article: PMC3099662] [PubMed: 21454587]
- 20.
- Kocher O, Krieger M. Role of the adaptor protein PDZK1 in controlling the HDL receptor SR-BI. Curr Opin Lipidol. 2009 Jun;20(3):236–41. [PMC free article: PMC2849150] [PubMed: 19421056]
Appendix A. Assay Summary Table
Table A1Summary of Completed Assays and AIDs
| PubChem AID | Type | Target | Concentration Range (µM) | Samples Tested |
|---|---|---|---|---|
| 488952 | Summary | SR-BI Inhibitor Project | NA | NA |
| 488896 | Cell-based | SR-BI, DiI-HDL uptake | 7.5 µM | 319,533 |
| 493194 | Cell-based | SR-BI, DiI-HDL uptake | 0.015-35 µM | 472 |
| 540354 | Cell-based | SR-BI, DiI-HDL uptake | 0.046-35 µM | 11 |
| 588392 | Cell-based | SR-BI, DiI-HDL uptake | 0.000195-35 µM | 31 |
| 588553 | Cell-based | SR-BI, DiI-HDL uptake | 0.015-35 µM | 32 |
| 588548 | Cell-based | SR-BI, DiI-HDL uptake | 0.015-35 µM | 32 |
| 588754 | Cell-based | SR-BI, DiI-HDL uptake | 28.5 fM-35 µM | 35 |
| 588828 | Cell-based | SR-BI, DiI-HDL uptake | 28.5 fM-35 µM | 35 |
| 588833 | Cell-based | SR-BI, DiI-HDL uptake | 28.5 fM-35 µM | 3 |
| 540246 | Cell-based | 24h Cytotoxicity | 0.0135- 35 µM | 66 |
| 588777 | Cell-based | SR-BI Alexa 488 HDL binding assay | 0.0135- 35 µM | 66 |
| 588403 | Biochemical | Fluorescence quencher assay | 0.000195-35 µM | 31 |
| 540326 | Biochemical | Fluorescence quencher assay | 0.015-35 µM | 472 |
| 588810 | Cell-based | SR-BI Alexa 488 HDL binding assay | 28.5 fM-35 µM | 35 |
| 588830 | Cell-based | 3h Cytotoxicity assay | 0.0135- 35 µM | 66 |
| 588829 | Cell-based | 24h Cytotoxicity assay | 28.5 fM-35 µM | 17 |
| 588826 | Cell-based | 24h Cytotoxicity assay | 28.5 fM-35 µM | 37 |
| 588825 | Cell-based | [ldlA7] DiI-HDL Counterscreen | 28.5 fM-35 µM | 17 |
| 588818 | Cell-based | Cholesterol efflux, wild type | 1-25 µM | 4 |
| 602152 | Cell-based | Cholesterol efflux, wild type | 0.001- 25 µM | 5 |
| 588843 | Cell-based | Cholesterol efflux, C384S mutant | 1-25 µM | 4 |
| 602138 | Cell-based | Cholesterol efflux, C384S mutant | 0.001- 25 µM | 5 |
| 602134 | Cell-based | Transferrin Endocytosis | 28.5 fM-35 µM | 3 |
| 588836 | Cell-based | Radiolabeled HDL uptake in WT cells | 0.0001-10 µM | 4 |
| 588837 | Cell-based | Radiolabeled HDL uptake in C384S mutant cells | 0.0001-10 µM | 4 |
| 602126 | Cell-based | 24h Cytotoxicity assay | 28.5 fM-35 µM | 138 |
| 588831 | Cell-based | DiI-HDL Uptake Assay for Reversibility | 28.5 fM-35 µM | 3 |
| 602154 | Cell-based | DiI-HDL Uptake Assay for Reversibility with extended washout | 28.5 fM-35 µM | 3 |
| 602155 | Biochemical | Radiolabeled HDL uptake with SR-BI in purified liposomes | 0.1-1 µM | 4 |
NA= not applicable
Appendix B. Detailed Assay Protocols
DiI-HDL Uptake Primary Assay (2085-01)
DiI-HDL Labeling Mix
- 2200 ml of Ham's/0.5% BSA/25 mM HEPES pH 7.4
- DiI-HDL to 10 µg protein/ml final concentration (Stocks vary from 2.81-4 mg/ml)
Ham's/0.5% BSA/25 mM HEPES Assay Media (1 liter)
- 5 g BSA powder (Sigma fatty acid-free)
- 1 Liter Ham's F12 base media
- 25 ml 1 M HEPES pH 7.4
ldlA[mSR-BI] Chinese Hamster Ovary (CHO) cells were maintained in Ham's F12K (Cellgro Catalog No. 10-080-CV)/10% Fetal Calf Serum (Hyclone Catalog No. SH30071.03, Lot No. ATG32533)/ 1x Penicillin-streptomycin-L-Glutamine (Gibco Catalog No.10378-016)/200 µg/mL Geniticin (Gibco Catalog No.10131-027, Lot No.802967). Cells were fluid changed every 2 days and/or split upon reaching 80% confluency. For the primary HTS, cells were thawed at 6 million cells per Falcon T225 flask. After 2 days, the cells were passed to a Corning Hyperflask and plated after 3 days in the Hyperflask with a fluid change the day prior to plating.
On Day 0
- Plate 10,000 ldlA[mSR-BI] cells in 30 µl per well in Ham's F12K (Cellgro Catalog No. 10-080-CV)/5% Fetal Calf Serum (Hyclone Catalog No. SH30071.03, Lot No. ATG32533)/ 1x Penicillin-streptomycin-L-Glutamine (Gibco Catalog No. 10378-016) with a Thermo Combi Multi-drop fluid handler.
- Use Aurora black 384-well, square, clear-bottomed, image-quality plates (Aurora Catalog No. 1022-11330) for the assay.
On Day 1
- Remove media with aspirator (Bio-Tek ELX405 plate washer).
- Add 30 µl Ham's F12/0.5% Bovine Serum Albumin (fatty acid-free) /25 mM HEPES pH 7.4 (Invitrogen) + 10 µg protein/ml DiI-HDL with Thermo Combi fluid handler (slow speed setting).
- Pin transfer 100 nl compounds and positive control (1 µM BLT-1).
- Incubate 3 hours @ 37 °C in humidified cell culture incubator.
- Remove media with aspirator.
- Wash twice with 30 µl PBS (+Ca and Mg) using the Thermo Combi on slow speed setting.
- Analyze DiI-HDL uptake with Perkin-Elmer EnVision plate reader (Bodipy TMR mirror #405, Excitation filter is Photometric 550 (#312) and emission filter is Cy3 595 (#229) with bottom read.
Cytotoxicity Assay (2085-02)
On Day 0
- Plate 4,000 cells [ldlA]mSR-BI 30 µl/well in Ham's F-12/10% fetal bovine serum (Hyclone)/Penicillin-Streptomycin-L-Glutamine.
On Day 1
- Pin transfer 100 nl compounds and positive control (sentinel plate).
- Incubate 3 hours @ 37 °C.
- Add 20 µl Promega CellTiter-Glo per well.
- Shake plate for 15 seconds.
- Incubate 10 minutes.
- Read luminescence with Perkin-Elmer EnVision plate reader.
HDL Binding Assay (2085-03)
ldlA[mSR-BI] Chinese Hamster Ovary (CHO) cells were maintained in Ham's F12K (Cellgro Catalog No. 10-080-CV)/10% Fetal Calf Serum (Hyclone Catalog No. SH30071.03, Lot No. ATG32533)/ 1x Penicillin-streptomycin-L-Glutamine (Gibco 10378-016)/200 µg/ml Geniticin (Gibco Catalog No.10131-027, Lot No.802967). Cells were fluid changed every 2 days and/or split upon reaching 80% confluency.
On Day 0
- Plate 10,000 ldlA[mSR-BI] cells in 30 µl per well in Ham's F12K (Cellgro Catalog No. 10-080-CV)/5% Fetal Calf Serum (Hyclone Catalog No. SH30071.03, Lot No. ATG32533)/ 1x Penicillin-streptomycin-L-Glutamine (Gibco 10378-016).
- Use Aurora black, 384-well, square, clear-bottomed image-quality plates (Aurora Catalog No.1022-11330) for the assay.
On Day 1
- Remove media with aspirator (ELX405 plate washer).
- Add 30 µl Ham's F12/0.5% Bovine Serum Albumin (fatty acid-free) (Sigma Catalog No. A8806-5G) /25 mM HEPES pH 7.4 (Invitrogen) plus 10 µg/ml Alexa 488-HDL with Thermo Combi fluid handler (slow speed setting).
- Pin transfer 100 nl compounds and positive control (1 µM BLT-1).
- Incubate 3 hours @ 37 °C in a humidified cell culture incubator.
- Remove media with aspirator.
- Wash twice with 30 µl PBS (+Ca and Mg) using the Thermo Combi on slow speed setting.
- Analyze Alexa-488-HDL binding with Perkin-Elmer EnVision plate reader (FITC mirror #403) with bottom read
DiI-HDL Uptake Counter Assay (2085-07)
DiI-HDL Labeling Mix
- 2200 ml of Ham's/0.5% BSA/25 mM HEPES pH 7.4
- DiI-HDL to 10 µg/ml final concentration (Stocks vary from 2.81-4 mg/ml)
Ham's/0.5% BSA/25 mM HEPES Assay Media (1 liter)
- 5 g BSA powder (Sigma fatty acid-free)
- 1 Liter Ham's F12 base media
- 25 ml 1 M HEPES pH 7.4
[ldlA7] Chinese Hamster Ovary (CHO) cells were maintained in Ham's F12K (Cellgro Catalog No. 10-080-CV)/10% Fetal Calf Serum (Hyclone Catalog No. SH30071.03, Lot No. ATG32533)/ 1x Penicillin-streptomycin-L-Glutamine (Gibco Catalog No.10378-016). Cells were fluid changed every 2 days and/or split upon reaching 80% confluency. For the primary HTS, cells were thawed at 6 million cells per Falcon T225 flask. After 2 days, the cells were passed to a Corning Hyperflask and plated after 3 days in the Hyperflask with a fluid change the day prior to plating.
On Day 0
- Plate 10,000 [ldlA7] cells in 30 µl per well in Ham's F12K (Cellgro Catalog No. 10-080-CV)/5% Fetal Calf Serum (Hyclone Catalog No. SH30071.03, Lot No. ATG32533)/ 1x Penicillin-streptomycin-L-Glutamine (Gibco Catalog No. 10378-016).
- Use Aurora black 384-well, square, clear-bottomed, image-quality plates (Aurora Catalog No. 1022-11330) for the assay.
On Day 1
- Remove media with aspirator (ELX405 plate washer).
- Add 30 µl Ham's F12/0.5% Bovine Serum Albumin (fatty acid-free) /25 mM HEPES pH 7.4 (Invitrogen) + 10 µg protein/ml DiI-HDL with Thermo Combi fluid handler (slow speed setting).
- Pin transfer 100 nl compounds and positive control (1 µM BLT-1).
- Incubate 3 hours @ 37 °C in humidified cell culture incubator.
- Remove media with aspirator.
- Wash twice with 30 µl PBS (+Ca and Mg) using the Thermo Combi on slow speed setting.
- Analyze DiI-HDL uptake with Perkin-Elmer EnVision plate reader (Bodipy TMR mirror #405, Excitation filter is Photometric 550 (#312) and emission filter is Cy3 595 (#229) with bottom read.
Cholesterol Efflux Assay (wild type [2085-06]) and C384S Mutant SR-BI (2085-10)
On Day 0
- Seed cells onto 24-well tissue culture plates at a density of 50,000 cells per well in 1 ml Ham's F-12/10% fetal bovine serum (Hyclone)/Penicillin-Streptomycin-L-Glutamine medium with G418/Geniticin at 250 µg/ml.
On Day 1
- Replace the medium with Ham's F12 medium supplemented with 10% bovine lipoprotein deficient serum, 1 µCi/ml [1,2-3H]cholesterol (40–60 Ci/mmol; Product number: NET139001MC Perkin Elmer - NEN Life Science), 2 mM L-glutamine, 50 U/ml penicillin, 50 mg/ml streptomycin, and, for ldlA[mSR-BI] cells only, 0.25 mg/ml G418.
On Day 3
- Washthe cells twice in 1 ml Ham's F12 medium without any supplements.
- Culture for another 24 h in 1 ml tissue culture medium in which fetal bovine serum was replaced with 1% fatty acid-free (FAF) BSA (Sigma Catalog No. A6003).
On Day 4
- Wash cells twice with Ham's F12 without supplements.
- Pre-incubate for 1 h at 37 degrees Celsius with compounds at several concentrations in 1 ml assay medium (Ham's F12 + P/S + Gln + 0.5% DMSO + 25 mM HEPES, pH 7.4+ 0.5% FAF BSA).
- Incubatethe cells for an additional 2 hours with the same concentrations of small molecules and with the indicated concentrations unlabeled HDL → 55 µl of assay medium containing 2 mg/ml HDL added to each well (final HDL concentration of 100 µg/ml).
- After 2hours of incubation at 37 °C, determine the [3H]cholesterol contents of the cells and the media as follows.
- Collect 120 ul efflux media and clarify by centrifugation for 1 min with a desktop microcentrifuge,
- Determine the radioactivity in 100 µl of each supernatant by liquid scintillation counting (LSC).
- Solubilize cells with 400 µl of lysis buffer (1% Triton X-100 in PBS) for 30 min at room temperature,
- Determine the amount of [3H]cholesterol in 100 µl of each lysate by LSC.
- Calculate the total cellular [3H]cholesterol as the sum of the radioactivity in the efflux medium plus the radioactivity in the cells. Use total cellular [3H]cholesterol to calculate the [3H]cholesterol efflux (percent of total [3H]cholesterol released into the medium).
DiI-HDL Uptake Reversibility Assay (2085-14)
DiI-HDL Labeling Mix
- 2200 ml of Ham's/0.5% BSA/25 mM HEPES
- DiI-HDL to 10 µg protein/ml final concentration (Stocks vary from 2.81-4 mg/ml)
Ham's/0.5% BSA/25 mM HEPES Assay Media (1 liter)
- 5 g BSA powder (Sigma fatty acid-free)
- 1 Liter Ham's F12 base media
- 25 ml 1 M HEPES pH 7.4
[ldlA7][mSR-BI] Chinese Hamster Ovary (CHO) cells were maintained in Ham's F12K (Cellgro Catalog No. 10-080-CV)/10% Fetal Calf Serum (Hyclone Catalog No. SH30071.03, Lot No. ATG32533)/ 1x Penicillin-streptomycin-L-Glutamine (Gibco Catalog No.10378-016)/ Geniticin 200 µg/ml. Cells were fluid changed every 2 days and/or split upon reaching 80% confluency.
On Day 0
- Plate 10,000 [ldlA]mSR-BI cells in 30 µl per well in Ham's F12K (Cellgro Catalog No. 10-080-CV)/5% Fetal Calf Serum (Hyclone Catalog No. SH30071.03, Lot No. ATG32533)/ 1x Penicillin-streptomycin-L-Glutamine (Gibco Catalog No. 10378-016).
- Use Aurora black 384-well, square, clear-bottomed, image-quality plates (Aurora Catalog No. 1022-11330) for the assay.
On Day 1
- Remove media with aspirator (ELX405 plate washer).
- Add 30 µl Ham's F12/0.5% Bovine Serum Albumin (fatty acid-free) /25 mM HEPES pH 7.4 (Invitrogen) with Thermo Combi fluid handler (slow speed setting).
- Pin transfer 100 nl compounds and positive control (1 µM BLT-1).
- Incubate 2 hours @ 37 °C in humidified cell culture incubator.
- Remove media with aspirator.
- Wash four times with 30 µl PBS (+Ca and Mg) using the Thermo Combi on slow speed setting.
- Add 30 µl Ham's F12/0.5% Bovine Serum Albumin (fatty acid-free) /25 mM HEPES pH 7.4 (Invitrogen) + 10 µg protein/ml DiI-HDL with Thermo Combi fluid handler (slow speed setting).
- Analyze DiI-HDL uptake with Perkin-Elmer EnVision plate reader (Bodipy TMR mirror #405, Excitation filter is Photometric 550 (#312) and emission filter is Cy3 595 (#229) with bottom read.
DiI-HDL Uptake Extended Reversibility Assay (2085-15)
DiI-HDL Labeling Mix
- 2200 ml of Ham's/0.5% BSA/25 mM HEPES
- DiI-HDL to 10 µg protein/ml final concentration (Stocks vary from 2.81-4 mg/ml)
Ham's/0.5% BSA/25 mM HEPES Assay Media (1 liter)
- 5 g BSA powder (Sigma fatty acid-free)
- 1 Liter Ham's F12 base media
- 25 ml 1 M HEPES
[ldlA7]]mSR-BI Chinese Hamster Ovary (CHO) cells were maintained in Ham's F12K (Cellgro Catalog No. 10-080-CV)/10% Fetal Calf Serum (Hyclone Catalog No. SH30071.03, Lot No. ATG32533)/ 1x Penicillin-streptomycin-L-Glutamine (Gibco Catalog No.10378-016)/200 µg/ml. Cells were fluid changed every 2 days and/or split upon reaching 80% confluency.
On Day 0
- Plate 10,000 [ldlA7] cells in 30 µl per well in Ham's F12K (Cellgro Catalog No. 10-080-CV)/5% Fetal Calf Serum (Hyclone Catalog No. SH30071.03, Lot No. ATG32533)/ 1x Penicillin-streptomycin-L-Glutamine (Gibco Catalog No. 10378-016).
- Use Aurora black 384-well, square, clear-bottomed, image-quality plates (Aurora Catalog No. 1022-11330) for the assay.
On Day 1
- Remove media with aspirator (ELX405 plate washer).
- Add 30 µl Ham's F12/0.5% Bovine Serum Albumin (fatty acid-free) /25 mM HEPES pH 7.4 (Invitrogen) with Thermo Combi fluid handler (slow speed setting).
- Pin transfer 100 nl compounds and positive control (1 µM BLT-1).
- Incubate 2 hours @ 37 °C in humidified cell culture incubator.
- Remove media with aspirator.
- Wash four times with 30 µl PBS (+Ca and Mg) using the Thermo Combi on slow speed setting.
- Add 30 µl Ham's F12/0.5% Bovine Serum Albumin (fatty acid-free) /25 mM HEPES pH 7.4 (Invitrogen) + 10 µg protein/ml DiI-HDL with Thermo Combi fluid handler (slow speed setting).
- Analyze DiI-HDL uptake with Perkin-Elmer EnVision plate reader (Bodipy TMR mirror #405, Excitation filter is Photometric 550 (#312) and emission filter is Cy3 595 (#229) with bottom read.
Transferrin Endocytosis Assay (2085-04)
On Day 0
- Plate 5,000 ldlA[mSR-BI] cells in 30 µl per well of a 384 well plate in DMEM/5% FCS/Pen-Strep.
On Day 1
- Wash 2x with 30 µl DMEM/1% BSA/(no phenol red).
- Dispense 30 µl per well of DMEM/1%BSA/no phenol red.
- Add compounds, incubate 60 min @ 37 °C.
- Dissolve 5 mg Transferrin-Alexa 594 (Invitrogen T-13343) in 1 ml water.
- Dilute 5 mg/ml stock 1:10 to become 5 µg/µl stock, add 1.8 µl per well for 30 µl volume (need approx. 750 µL per 384-well plate).
- Incubate for 30 min @ 37 °C.
- Place plate on ice to stop reaction.
- Wash wells with 45 µl ice cold PBS/1 mM MgCl2/0.1 mM CaCl2.
- Fix for 45 min on ice with ice cold 4% PFA in PBS/1 mM MgCl2/0.1mM CaCl2 in the presence of DAPI (1:1000 from a 10 mg/ml stock; Invitrogen No.H3570).
- Analyze with high content imaging.
Liposome Uptake Assay (2085-16)
Liposome prep
Set up
- 10 mg phosphatidyl Choline (PC) (purchased at 10mg/ml, so add 1 ml)
- 1 mg choline (C) (dissolve in 10 µl CH3Cl chloroform to make it 10 mg/ml)
- Add into a glass scintillation vial.
- Dry with liquid N2 and rotate the vial to dry the lipids.
- Reconstitute in 5 ml 50 mM Tris pH 6.0 buffer:
- Add 2 ml first, vortex, and shake on a platform for 5 min, transfer to a new vial.
- Add another 2 ml to the original vial and repeat the same step.
- Add 1 ml to clean the vial and transfer all lipids into the new vial.
Liposome can be stored at 4 ºC for 1 week, or 7-12 days.
Proteoliposome prep
Make fresh proteoliposome prep for each experiment, and also make one without SR-BI.
SR-BI protein sample
- Thaw and put on ice.
- Spin 15 min @ 14,000 RPM in cold room.
- Take the supernatant.
Label Oak Ridge centrifuge tubes and place on ice.
For each tube
- Add 2.4 ml buffer (50 mM Tris-Cl, pH 6.0, 150 mM NaCl, 2 mM CaCl2).
- Add 20 µg SR-BI or buffer. Vortex.
- Add PC-C liposome: 0.5 mL. Vortex.
- Add acetone (0.6x volume, ice cold). Vortex.
- Centrifuge (ultracentrifuge, red rotor, Ti 70.1) @ 16,000 RPM, 30 min, 4 ºC.
- Aspirate supernatant, dry upside down for 5 min.
- Add 375 µl cold Ham's F12 + 25 mM HEPES. Vortex.
- Sit on ice for 5 min.
- Add another 750 µl of Ham's F12 + 25 mM HEPES.
- Transfer into microcentrifuge tubes.
- 1125 µl per tube(30 µl proteoliposome is used per sample, so each prep is OK for maximum of 37 samples).
Uptake/binding Assay
Final assay condition in Ham's F12 + 25 mM HEPES + 0.5% DMSO + 0.5% FAFA
In 96-well plate,
- Add:
- -
30 μl liposome in F12 + 25 mM HEPES.
- -
30 μl BLT-1 or other compounds or cold HDL in Ham's F12 + 25 mM HEPES + 1% DMSO + 1% FAFA (prepared 2.67X concentrated).
- -
20 μl ‘radiolabeled HDL in Ham's F12 + 25 mM HEPES + 0.5% DMSO + 0.5% FAFA (prepared 4X concentrated).
- Mix by pipetting up and down 2-3 times.
- After 1 hour incubation at 37 ºC, add radiolabeled HDL for 4 hours.
- To stop the reaction, put the plate on ice for 10 min – keep the plate on ice through filter assay.
- Proceed to Filtration.
Filter Assay
- Prewash/block filters for > 60 min @ RT in wash buffer (20 mM Tris-Cl pH 8.0, 50 mM NaCl, 20 µM CaCl2, 1 mg/mL BSA).
- Mount membrane in filtration bucket.
- Add 200 µl washing buffer to the plates: 12 samples at a time.Apply samples (~ 300 µl) to filter while vacuum is ON.
- Add/wash wells with another 200 µl washing buffer.Apply samples to filter while vacuum is ON.
- Vacuum OFF, add 6 ml wash buffer to filter → vacuum ON to suck.
- Dry filters as much as possible, and remove the filter.
- For 3H, add 4 ml scintillation liquid and wait until the filters are dissolved and count. For 125I: make sure membrane is crammed within tip of tube.
Data Analysis
For the primary screen and other assays, negative-control (NC) wells and positive-control (PC) wells were included on every plate. The raw signals of the plate wells were normalized using the ‘Stimulators Minus Neutral Controls’ method in Genedata Assay Analyzer (v7.0.3). The median raw signal of the intra-plate NC wells was set to a normalized activity value of 0, while the median raw signal of the intra-plate PC wells was set to a normalized activity value of 100. Experimental wells were scaled to this range, resulting in an activity score representing the percent change in signal relative to the intra-plate controls. The mean of the replicate percent activities were presented as the final ‘Pubchem Activity Score’.
The ‘Pubchem Activity Outcome’ class was assigned as described below, based on an activity threshold of 70%:
- Activity_Outcome = 1 (inactive), less than half of the replicates fell outside the threshold.
- Activity_Outcome = 2 (active), all of the replicates fell outside the threshold, OR at least half of the replicates fell outside the threshold AND the ‘Pubchem Activity Score’ fell outside the threshold.
- Activity_Outcome = 3 (inconclusive), at least half of the replicates fell outside the threshold AND the ‘Pubchem Activity Score did not fall outside the threshold.
Appendix C. Experimental Procedures for the Synthesis of the Probes
General Details. All reagents and solvents were purchased from commercial vendors and used as received. NMR spectra were recorded on a Bruker 300 MHz or Varian UNITY INOVA 500 MHz spectrometer as indicated. Proton and carbon chemical shifts are reported in parts per million (ppm;δ) relative to tetramethylsilane, CDCl3 solvent, or d6-DMSO (1H δ 0, 13C δ 77.16, or 13C δ 39.5, respectively). NMR data are reported as follows: chemical shifts, multiplicity (obs = obscured, app = apparent, br = broad, s = singlet, d = doublet, t = triplet, q = quartet, p = pentet, sext = sextet, sept = septet, m = multiplet, comp = complex overlapping signals); coupling constant(s) in Hz; integration. Unless otherwise indicated, NMR data were collected at 25°C. Flash chromatography was performed using 40-60 µm Silica Gel (60 Å mesh) on a Teledyne Isco Combiflash Rf system. Tandem liquid chromatography/mass spectrometry (LCMS) was performed on a Waters 2795 separations module and Waters 3100 mass detector. Analytical thin layer chromatography (TLC) was performed on EM Reagent 0.25 mm silica gel 60-F plates. Visualization was accomplished with UV light and aqueous potassium permanganate (KMnO4) stain followed by heating. High-resolution mass spectra were obtained at the MIT Mass Spectrometry Facility with a Bruker Daltonics APEXIV 4.7 Tesla Fourier Transform Ion Cyclotron Resonance mass spectrometer. Compound purity and identity were determined by UPLC-MS (Waters, Milford, MA). Purity was measured by UV absorbance at 210 nm. Identity was determined on a SQ mass spectrometer by positive electrospray ionization. Mobile Phase A consisted of either 0.1% ammonium hydroxide or 0.1% trifluoroacetic acid in water, while mobile Phase B consisted of the same additives in acetonitrile. The gradient ran from 5% to 95% mobile Phase B over 0.8 minutes at 0.45 mL/min. An Acquity BEH C18, 1.7 µm, 1.0 ×50 mm column was used with column temperature maintained at 65°C. Compounds were dissolved in DMSO at a nominal concentration of 1 mg/mL, and 0.25 µL of this solution was injected.
5-(Benzo[d][1,3]dioxol-5-yl)-2H-tetrazole: A round-bottom flask equipped with a magnetic stirbar was charged with benzo[d][1,3]dioxole-5-carbonitrile (2.50 g, 17.0 mmol) and N,N-dimethylformamide (17.0 mL). Sodium azide (1.22 g, 18.7 mmol) and ammonium chloride (1.00 g, 18.7 mmol) were then added to give a white suspension. This mixture was heated to 150 °C and stirred for 24 hours. The reaction was cooled to ~70 °C, then poured on ice (~50 mL). The mixture was acidified to pH 1 with 1.0 M aqueous hydrochloric acid solution. The precipitated solids were collected by filtration, washed with ice-cold water (10 mL), and air-dried on the filter to give the title compound as a white solid (2.79 g, 86%). 1H NMR (300 MHz, CD3OD): δ 7.55 (dd, J = 8.1, 1.7 Hz, 1H), 7.46 (d, J = 1.5 Hz, 1H), 7.02 (d, J = 8.1 Hz, 1H), 6.08 (s, 2H); MS (ESI+): 191 (M+H).

Methyl 2-(5-(benzo[d][1,3]dioxol-5-yl)-2H-tetrazol-2-yl)acetate: A round-bottom flask equipped with a magnetic stirbar was charged with 5-(benzo[d][1,3]dioxol-5-yl)-2H-tetrazole (2.79 g, 14.7 mmol) and acetone (49.0 mL). The suspension was cooled to 0 °C before adding anhydrous potassium carbonate. After stirring for 30 minutes at 0 °C, methyl bromoacetate (1.5 mL, 16.2 mmol) was added to the opaque, tan mixture. The reaction was stirred for 18 hours while slowly warming to room temperature. The mixture was filtered through a pad of Celite ®, and the filter was thoroughly washed with additional acetone. The clear, yellow filtrate was concentrated under reduced pressure to give an orange solid. This mixture of regioisomers (~8:1 ratio) was separated by column chromatography over silica gel (hexanes/ethyl acetate: 100/0 to 40/60) to give the title compound as a white solid (2.93 g, 76 %). 1H NMR (300 MHz, CDCl3): δ 7.71 (dd, J = 8.1, 1.6 Hz, 1H), 7.59 (d, J = 1.5 Hz, 1H), 6.91 (d, J = 8.1 Hz, 1H), 6.03 (s, 2H), 5.45 (s, 2H), 3.82 (s, 3H); MS (ESI+): 263 (M+H).

2-(5-(Benzo[d][1,3]dioxol-5-yl)-2H-tetrazol-2-yl)acetic acid: Methyl 2-(5-(benzo[d][1,3]dioxol-5-yl)-2H-tetrazol-2-yl)acetate (2.93 g, 11.2 mmol) was placed in a round-bottom flask equipped with a magnetic stirbar and dissolved with dichloromethane (33.5 mL) and methanol (3.7 mL). Sodium hydroxide pellets (0.89 g, 22.3 mmol) were added to the clear, pale yellow solution. The reaction was stirred at room temperature for 1 hour and the resulting opaque, white mixture was concentrated under reduced pressure to give a white solid. This material was dissolved in water (250 mL) and washed with dichloromethane (2 ×100 mL). The aqueous phase was acidified to pH 1 with concentrated hydrochloric acid to give an opaque white mixture. Extraction with hot ethyl acetate (3 ×100 mL) produced a clear aqueous layer. The combined ethyl acetate extracts were shaken over magnesium sulfate, filtered, and concentrated under reduced pressure to give title compound as a white solid (2.64 g, 95 %). 1H NMR (300 MHz, d6-DMSO): δ 7.62 (d, J = 8.0 Hz, 1H), 7.52 (s, 1H), 7.11 (d, J = 7.9 Hz, 1H), 6.14 (s, 2H), 5.70 (s, 2H); MS (ESI+): 249 (M+H).
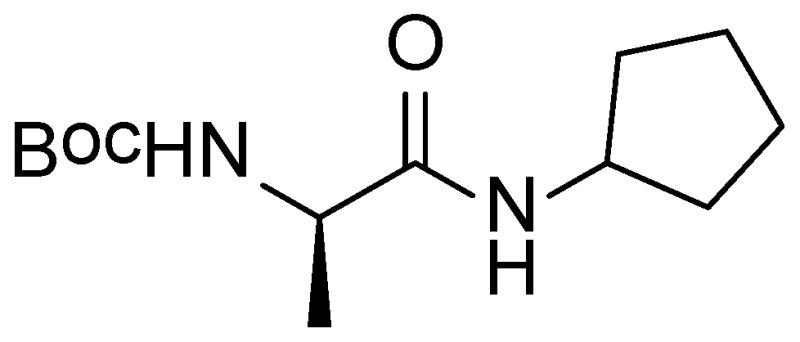
(R)-tert-butyl (1-(cyclopentylamino)-1-oxopropan-2-yl)carbamate: N-Boc-D-alanine (250 mg, 1.32 mmol) and HOBt (243 mg, 1.59 mmol) were added to a vial with stir bar and dissolved with dry DCM (6.6 mL)). Hunig's base (254 µL, 1.45 mmol) was then added, followed by cyclopentylamine (124 mg, 1.45 mmol), and EDC (304 mg, 1.59 mmol). The reaction was then stirred for 18 h, after which time LC-MS showed complete consumption of starting material and conversion to the desired product. The reaction was diluted with 1 M aq. HCl and EtOAc, then the phases were separated, and the combined organics were washed again with aq. HCl, then twice with half-saturated aq. NaHCO3, then brine. The organics were dried over Na2SO4, filtered, and concentrated to yield the title compound as a white solid (311 mg, 92%). 1H NMR (300 MHz, CDCl3): δ 6.11 (br s, 1H), 5.00 (br s, 1H), 4.19 (app sext, J = 6.6 Hz, 1H), 4.08 (m, 1H), 1.93 (m, 2H), 1.61 (m, 4H), 1.47 (s, 9H), 1.39 (m, 2H), 1.32 (t, J = 8.0 Hz, 3H); MS (ESI+): 257.37 (M+H).
(R)-2-(cyclohexylamino)-N-cyclopentylpropanamide: (R)-tert-butyl 1-(cyclopentylamino)-1-oxopropan-2-ylcarbamate (310 mg, 1.21 mmol), DCM (6.0 mL), triethylsilane (0.97 mL, 6.1 mmol), and TFA (0.93 mL, 12.1 mmol) were added to a flask with stir bar and stirred for 18 h. The reaction was then quenched by pipetting the reaction solution slowly into a stirred half-saturated aqueous solution of NaHCO3 (50 mL). The mixture was diluted with DCM, the layers were separated, and the organics were washed again with aq. NaHCO3, then brine. The organics were dried over Na2SO4, filtered, and concentrated, yielding the intermediate primary amine as an off-white solid. This was immediately dissolved with methanol (6.1 mL) and cooled on ice. Cyclohexanone (0.38 mL, 3.6 mmol) was added, followed by sodium cyanoborohydride (760 mg, 12.1 mmol) added portionwise over 10 min. The reaction was stirred for 24 h, after which time LC-MS analysis showed complete conversion to the desired product. The reaction was concentrated to remove most of the MeOH, then it was diluted with half-saturated aq. NaHCO3 and ether. The layers were separated, then the combined organics were washed again with NaHCO3. Dilute aq. HCl was then added to bring the product into the aqueous layer, which was then washed twice with ether to remove the excess cyclohexanone. The aqueous layer was basified again w/ aq. NaHCO3, then extracted twice with Et2O. The ether layer was then washed with brine and dried over Na2SO4, filtered, and concentrated to provide the title compound as a white solid (239 mg, 83% over two steps). A portion of the material was converted to the hydrochloride salt by dissolving in ether and adding excess HCl in Et2O, then concentrating. 1H NMR (300 MHz, CD3OD): δ 4.13 (app p, J = 6.7 Hz, 1H), 3.95 (q, J = 6.9 Hz, 1H), 2.97 (m, 1H), 2.15 – 1.80 (comp, 6H), 1.80 – 1.55 (comp, 6H), 1.48 (d, J = 6.9 Hz, 3H), 1.40 – 1.15 (comp, 6H); MS (ESI+): 239.68 (M+H).
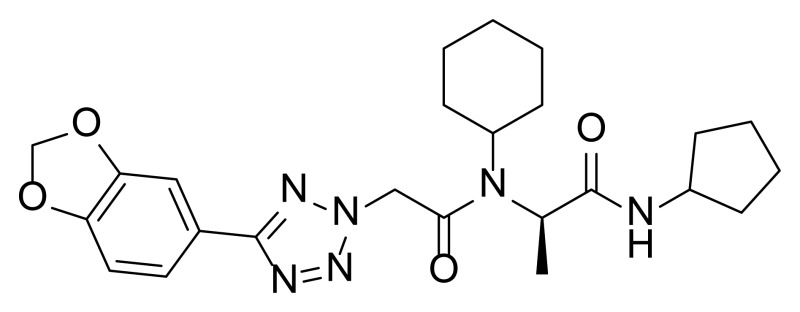
(R)-2-(2-(5-(benzo[d][1,3]dioxol-5-yl)-2H-tetrazol-2-yl)-N-cyclohexylacetamido)-N-cyclopentylpropanamide (PROBE): 2-(5-(benzo[d][1,3]dioxol-5-yl)-2H-tetrazol-2-yl)acetic acid (92 mg, 0.37 mmol) was sealed under nitrogen in a vial with stir bar. Dry DCM (2.2 mL), oxalyl chloride (32 µL, 0.37 mmol), and DMF (1 drop from a 22-gauge needle) were added, and the reaction was vented to an oil bubbler. The reaction was stirred for 4 h, then (R)-2-(cyclohexylamino)-N-cyclopentylpropanamide (free base, 80 mg, 0.34 mmol) was added to the resulting solution of acid chloride, followed by Hunig's Base (129 µl, 0.74 mmol) and DMAP (4.1 mg, 0.034 mmol). The reaction was stirred for 16 h, after which time LC-MS analysis showed complete conversion to the desired product. The reaction was diluted with 1 M aq. HCl and EtOAc (~25 mL each), the layers were separated, and the combined organics were washed again with aq. HCl, then twice with half-saturated aq. NaHCO3, then brine. The organic layer was dried over Na2SO4, filtered, and concentrated to a yellow oil, then purified by column chromatography (0 to 50% EtOAc/hexanes) to yield the title product as a pale yellow solid, after concentrating from an ether solution made cloudy by slowly adding pentane (90 mg, 72%). 1H NMR (300 MHz, CDCl3) δ 7.70 (dd, J = 8.1, 1.5 Hz, 1H), 7.58 (d, J = 1.4 Hz, 1H), 6.91 (d, J = 8.1 Hz, 1H), 6.60 (br d, J = 6.8 Hz, 1H), 6.04 (s, 2H), 5.59 (d, J = 15.4 Hz, 1 H), 5.48 (d, J = 15.4 Hz, 1H), 4.10 (m, 1H), 3.96 (br m, 1H), 3.52 (br app t, J = 11.4 Hz), 2.08 – 0.82 (comp, 21H) (peaks are broad due to rotamers); 13C NMR (75 MHz, CDCl3) δ 170.9, 165.4, 163.8, 149.6, 148.2, 121.6, 121.1, 108.8, 107.4, 101.6, 77.6, 77.2, 76.7, 58.9, 55.7, 55.1, 51.5, 33.0, 32.9, 31.6, 31.4, 25.9, 25.8, 25.0, 23.8, 15.9 (extra peaks are due to the minor rotamer). HRMS (ESI+): calculated for C24H33N6O4 [M+H] 469.2563, found 469.2552.
Appendix D. Experimental Procedure for Additional Analytical Assays
Solubility. Solubility was determined in phosphate buffered saline (PBS) pH 7.4 with 1% DMSO. Each compound was prepared in duplicate at 100 µM in both 100% DMSO and PBS with 1% DMSO. Compounds were allowed to equilibrate at room temperature with a 250 rpm orbital shake for 24 hours. After equilibration, samples were analyzed by UPLC-MS (Waters, Milford, MA) with compounds detected by SIR detection on a single quadrupole mass spectrometer. The DMSO samples were used to create a two-point calibration curve to which the response in PBS was fit.
PBS Stability. Stability was determined in the presence of PBS pH 7.4 with 0.1% DMSO. Each compound was prepared in duplicate on six separate plates and allowed to equilibrate at room temperature with a 250-rpm orbital shake for 48 hours. One plate was removed at each time point (0, 2, 4, 8, 24, and 48 hours). An aliquot was removed from each well and analyzed by UPLC-MS (Waters, Milford, MA) with compounds detected by SIR detection on a single quadrupole mass spectrometer. Additionally, to the remaining material at each time point, acetonitrile was added to force dissolution of compound (to test for recovery of compound). An aliquot of this was also analyzed by UPLC-MS.
GSH Stability. Stability was determined in the presence of PBS pH 7.4 µM and 50 µM glutathione with 0.1% DMSO. Each compound was prepared in duplicate on six separate plates and allowed to equilibrate at room temperature with a 250-rpm orbital shake for 48 hours. One plate was removed at each time point (0, 2, 4, 8, 24, and 48 hours). An aliquot was removed from each well and analyzed by UPLC-MS (Waters, Milford, MA) with compounds detected by SIR detection on a single quadrupole mass spectrometer. Additionally, to the remaining material at each time point, acetonitrile was added to force dissolution of compound (to test for recovery of compound). An aliquot of this was also analyzed by UPLC-MS.
Plasma Protein Binding. Plasma protein binding was determined by equilibrium dialysis using the Rapid Equilibrium Dialysis (RED) device (Pierce Biotechnology, Rockford, IL) for both human and mouse plasma. Each compound was prepared in duplicate at 5 µM in plasma (0.95% acetonitrile, 0.05% DMSO) and added to one side of the membrane (200 µl) with PBS pH 7.4 added to the other side (350 µl). Compounds were incubated at 37 ºC for 5 hours with a 250-rpm orbital shake. After incubation, samples were analyzed by UPLC-MS (Waters, Milford, MA) with compounds detected by SIR detection on a single quadrupole mass spectrometer.
Plasma Stability. Plasma stability was determined at 37 ºC at 5 hours in both human and mouse plasma. Each compound was prepared in duplicate at 5 µM in plasma diluted 50/50 (v/v) with PBS pH 7.4 (0.95% acetonitrile, 0.05% DMSO). Compounds were incubated at 37 ºC for 5 hours with a 250-rpm orbital shake with time points taken at 0 hours and 5 hours. Samples were analyzed by UPLC-MS (Waters, Milford, MA) with compounds detected by SIR detection on a single quadrupole mass spectrometer.
Appendix E. Chemical Characterization Data for Probe 2
13C NMR Spectrum (75 MHz, CDCl3) of Probe 2 (ML279)
Extra peaks in alkyl region due to a minor rotamer.
Appendix F. Prior Art Search
Appendix G. Compounds Provided to Evotec
Table A3Probe and Analog Information
| BRD | SID | CID | P/A | MLSID | ML |
|---|---|---|---|---|---|
| BRD-K92916754-001-03-3 | 126700838 | 53393835 | P | MLS003875059 | ML279 |
| BRD-A18255019-001-02-2 | 126922000 | 53262912 | A | MLS003875068 | NA |
| BRD-A90568062-001-02-5 | 126922004 | 53347965 | A | MLS003875070 | NA |
| BRD-A53666875-001-02-9 | 126922002 | 53377445 | A | MLS003875067 | NA |
| BRD-A87455084-001-02-4 | 126922003 | 53377432 | A | MLS003875066 | NA |
| BRD-K69535827-001-02-5 | 126922009 | 53377422 | A | MLS003875061 | NA |
A = analog; NA= not applicable; P = probe
Appendix H. Additional Analogs
Table A4SAR analysis of Additional Analogs
Appendix I. Chemical Characterization Data for All Analogs
- Probe Structure and Characteristics
- Recommendations for scientific use of the probe
- Introduction
- Materials and Methods
- Results
- Discussion
- References
- Assay Summary Table
- Detailed Assay Protocols
- Experimental Procedures for the Synthesis of the Probes
- Experimental Procedure for Additional Analytical Assays
- Chemical Characterization Data for Probe 2
- Prior Art Search
- Compounds Provided to Evotec
- Additional Analogs
- Chemical Characterization Data for All Analogs
- PMCPubMed Central citations
- PubChem BioAssay for Chemical ProbePubChem BioAssay records reporting screening data for the development of the chemical probe(s) described in this book chapter
- PubChem SubstanceRelated PubChem Substances
- PubMedLinks to PubMed
- Review A Small Molecule Inhibitor of Scavenger Receptor BI-mediated Lipid Uptake—Probe 1.[Probe Reports from the NIH Mol...]Review A Small Molecule Inhibitor of Scavenger Receptor BI-mediated Lipid Uptake—Probe 1.Faloon PW, Dockendorff C, Youngsaye W, Yu M, Nag PP, Lewis TA, Bennion M, Paterson C, Lam G, Dandapani S, et al. Probe Reports from the NIH Molecular Libraries Program. 2010
- Review A Small Molecule Inhibitor of Scavenger Receptor BI-mediated Lipid Uptake - Probe 3.[Probe Reports from the NIH Mol...]Review A Small Molecule Inhibitor of Scavenger Receptor BI-mediated Lipid Uptake - Probe 3.Faloon PW, Dockendorff C, Yu M, Bennion M, Johnston S, Negri J, Dandapani S, Munoz B, Perez JR, Palmer M, et al. Probe Reports from the NIH Molecular Libraries Program. 2010
- Discovery of bisamide-heterocycles as inhibitors of scavenger receptor BI (SR-BI)-mediated lipid uptake.[Bioorg Med Chem Lett. 2015]Discovery of bisamide-heterocycles as inhibitors of scavenger receptor BI (SR-BI)-mediated lipid uptake.Dockendorff C, Faloon PW, Germain A, Yu M, Youngsaye W, Nag PP, Bennion M, Penman M, Nieland TJ, Dandapani S, et al. Bioorg Med Chem Lett. 2015 Jun 15; 25(12):2594-8. Epub 2015 Apr 11.
- Indolinyl-Thiazole Based Inhibitors of Scavenger Receptor-BI (SR-BI)-Mediated Lipid Transport.[ACS Med Chem Lett. 2015]Indolinyl-Thiazole Based Inhibitors of Scavenger Receptor-BI (SR-BI)-Mediated Lipid Transport.Dockendorff C, Faloon PW, Yu M, Youngsaye W, Penman M, Nieland TJ, Nag PP, Lewis TA, Pu J, Bennion M, et al. ACS Med Chem Lett. 2015 Apr 9; 6(4):375-380. Epub 2015 Feb 2.
- Discovery of chemical inhibitors of the selective transfer of lipids mediated by the HDL receptor SR-BI.[Proc Natl Acad Sci U S A. 2002]Discovery of chemical inhibitors of the selective transfer of lipids mediated by the HDL receptor SR-BI.Nieland TJ, Penman M, Dori L, Krieger M, Kirchhausen T. Proc Natl Acad Sci U S A. 2002 Nov 26; 99(24):15422-7. Epub 2002 Nov 18.
- A Small Molecule Inhibitor of Scavenger Receptor BI-mediated Lipid Uptake—Probe ...A Small Molecule Inhibitor of Scavenger Receptor BI-mediated Lipid Uptake—Probe 2 - Probe Reports from the NIH Molecular Libraries Program
Your browsing activity is empty.
Activity recording is turned off.
See more...