NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
Lipid droplets (LDs) are the universal lipid storage organelles [1, 2]. Lipids remobilized from LDs are used both for energy production via beta-oxidation or anabolic reactions, such as membrane biosynthesis [3–5]. We have previously described three probes ML206, ML219, and ML220, which were optimized from high-throughput screening hits of Drosophila melanogaster embryonic cells and showed a potent LD reduction phenotype, with EC50s of 8 nM, 2 nM, and 705 nM, respectively. Herein, we describe the further optimization of ML219 and characterization of the new probe, ML360. This compound showed potent inhibition of LD formation in the primary Drosophila melanogaster cell line as well as three mammalian cell lines (3T3-L1, COS-7, and AML12). It retained potent activity in 3T3-L1 pre-adipocyte to adipocyte differentiation conditions and showed no signs of cytotoxicity over this extended protocol.
Assigned Assay Grant #: MH085686
Screening Center Name & PI: NIH Chemical Genomics Center, Christopher P. Austin
Chemistry Center Name & PI: NIH Chemical Genomics Center, Christopher P. Austin
Assay Submitter & Institution: Mathias Beller, Max-Planck-Institute for Biophysical Chemistry.
PubChem Summary Bioassay Identifier (AID): 1623
Probe Structure & Characteristics
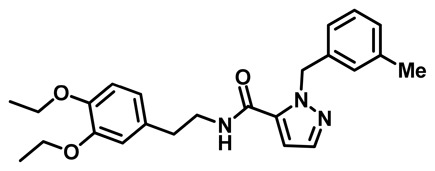
ML360
| CID/ML# | Target Name | IC50/EC50 (nM) [SID, AID] | Anti-target Name(s) | IC50/EC50 (μM) [SID, AID] | Fold Selective | Secondary Assay(s) Name: IC50/EC50 (nM) [SID, AID] |
|---|---|---|---|---|---|---|
| CID 70789742/ ML360 | 3T3L1 | 11 nM [SID 70789742, AID 652180] | Viability Assay | >20 μM [SID 70789742, AID 686935] | >1000-fold | Viability Assay [SID 70789742, AID 686935] |
1. Recommendations for Scientific Use of the Probe
ML360 showed a potent LD reduction phenotype in Drosophila melanogaster S3 cells, simian COS-7 cells, and murine 3T3-L1 and AML12 cells. While the target for this probe is still unknown, we have shown that this chemotype inhibits the formation of LDs rather than increase the lipolysis of established LDs. The potent activity ML360 in mammalian cells makes it a worthwhile tool for biologists to use in studying the formation and biological role of LDs.
2. Materials and Methods
General Chemistry Methods: All air or moisture sensitive reactions were performed under positive pressure of nitrogen with oven-dried glassware. Anhydrous solvents such as dichloromethane (DCM), N,N-dimethylforamide (DMF), acetonitrile, methanol, triethylamine, and trifluoroacetic acid (TFA) were purchased from Sigma-Aldrich. Preparative purification was performed on a Waters semi-preparative HPLC system. The column used was a Phenomenex Luna C18 (5 micron, 30 × 75 mm) at a flow rate of 45 mL/min. The mobile phase consisted of acetonitrile and water (each containing 0.1% trifluoroacetic acid). A gradient of 10% to 50% acetonitrile over 8 minutes was used during the purification. Fraction collection was triggered by UV detection (220 nM). Analytical analysis was performed on an Agilent LC/MS (Agilent Technologies, Santa Clara, CA). Method 1: A 7 minute gradient of 4% to 100% acetonitrile (containing 0.025% trifluoroacetic acid) in water (containing 0.05% trifluoroacetic acid) was used with an 8 minute run time at a flow rate of 1 mL/min. A Phenomenex Luna C18 column (3 micron, 3 × 75 mm) was used at a temperature of 50 °C. Method 2: A 3 minute gradient of 4% to 100% Acetonitrile (containing 0.025% trifluoroacetic acid) in water (containing 0.05% trifluoroacetic acid) was used with a 4.5 minute run time at a flow rate of 1 mL/min. A Phenomenex Gemini Phenyl column (3 micron, 3 × 100 mm) was used at a temperature of 50 °C. Purity determination was performed using an Agilent Diode Array Detector for both Method 1 and Method 2. Mass determination was performed using an Agilent 6130 mass spectrometer with electrospray ionization in the positive mode. 1H NMR spectra were recorded on Varian 400 MHz spectrometers. Chemical Shifts are reported in ppm with undeuterated solvent (DMSO-h6 at 2.49 ppm) as internal standard for DMSO-d6 solutions. All of the analogs tested in the biological assays have purity greater than 95% based on both analytical methods. High resolution mass spectrometry was recorded on Agilent 6210 Time-of-Flight LC/MS system. Confirmation of molecular formulae was accomplished using electrospray ionization in the positive mode with the Agilent Masshunter software (version B.02).
Regioisomeric Determination via Reverse Phase Chromatography. The regiochemistry was assigned by comparing the NOESY spectra of the two analogs. No correlation was seen between the pyrazole hydrogen (H) -3 proton and benzyl H-23 protons for ML360 while a strong NOESY correlation did exist between H-3 and H-23 for the regioisomer confirming the regiochemistry.
2.1. Assays
Lipid droplet (LD) accumulation in embryonicDrosophilaS3 Cells (AID 652175). The LD qHTS was performed with embryonic Drosophila S3 cells. Four μL of cells were dispensed into a LoBase Aurora COC 1,536-well plates to obtain a final concentration of 5,000 cells/well. Triacsin C, an inhibitor of long-chain fatty acyl CoA synthetase that blocks synthesis of triglycerides, was used as a positive control for lipid under-storage. A total of 23 nL of compounds dissolved in DMSO at different concentrations were transferred to the assay plates using a Kalypsys pin tool equipped with a 1,536-pin array. The final DMSO concentration was 0.46%. One μL of 400 μM oleic acid was added to induce lipid droplet formation in the cells, and the plates were lidded with stainless steel rubber gasket lined lids containing pinholes. After 18–24 hr incubation at 25 °C and 95% humidity, 4 μL of BODIPY lipid dye 493/503 and Cell Tracker Red CMTPX dye were added to the wells to stain lipid droplets and enumerate cell number, respectively. Fluorescence was detected by excitation of the fluorophores with a 488-nm laser on an Acumen Explorer. The total intensity in channel 1 (500–530 nm) reflected lipid droplet accumulation. Cells were detected using channel 3 (575–640 nm) with 5-micron width and 100-micron depth filters. The ratio of the total intensity in PMT channel 1 over total intensity of channel 3 was also calculated. Percent activity was computed relative to 100% inhibited lipid droplet deposition due to the presence of 20 μM Triacsin C.
Cell Viability Assay in mouse fibroblast-like pre-adipocyte 3T3-L1 cells (AID 686935). To assess toxicity of cherry picked compounds, a cellular viability assay was developed using the mouse adipocyte precursor 3T3-L1 cells. Four microliter of cells suspension in OPTI-MEM + 5% fetal bovine serum (FBS) media was dispensed into a 1536 well Poly-D-Lysine coated black imaging plates to obtain a final concentration of 500 cells/well. The assay plate was cultured at the incubation conditions 37 °C, 95% humidity and 5% CO2 overnight. Triacsin C was used as a positive control for lipid under-storage. A total of 23 nL of compounds dissolved in DMSO at different concentrations were transferred to the assay plates using a Kalypsys 1536-pin tool. The final DMSO concentration was 0.46%. One microliter of 400 μM oleic acid was added to induce lipid droplet formation in the cells, and the plates were lidded. After 18–24 hours of incubation, the cells were fixed with 4 % paraformaldehyde in PBS for 30 min at room temperature, washed three times with PBS, and stained for lipid droplet and nucleus using 4 μL of PBS solution containing Hoechst 33342. Images were acquired using InCell 2000 (GE) at 20× magnification. The number of nucleus was determined for cell viability by using GE Investigator software.
Lipid Droplet Accumulation in Mouse Fibroblast-like Pre-Adipocyte 3T3-L1 Cells (AID 652180). To evaluate cellular activity further and establish that the inhibitory effect of the compound is not cell line selective, an LD assay using mouse 3T3-L1 cells was developed. Four μL of cell suspension was dispensed into 1536 well Poly-D-Lysine coated black imaging plates (500 cells/well) and cultured at 37 °C, 95% humidity, and 5% CO2 overnight. Triacsin C was used as a positive control for lipid under-storage. A total of 23 nL of compounds dissolved in DMSO at different concentrations were transferred to the assay plates using a Kalypsys 1536-pin tool. The final DMSO concentration was 0.46%. One microliter of 400 μM oleic acid was added to induce lipid droplet formation in the cells. The plates were lidded and incubated at 37 °C, 95% humidity and 5% CO2 for 18–24 hours. The cells were then fixed with 4 % paraformaldehyde in PBS for 30 min at room temperature, washed three times with PBS and stained for lipid droplet and nucleus using 4 μL of PBS solution containing BODIPY lipid dye 493/503 and Hoechst 33342 respectively. Images were acquired using GE InCell 2000 at 20× magnification. The number of cells, number of lipids, and size of lipids were determined using GE Investigator software and histogram dose response plots were generated.
Differentiation of Mouse Fibroblast/Pre-Adipocyte Cells into Adipocyte Cells (AID: 652183). To assess the functional effect of the probe on lipid cell, a differentiation assay was developed using the 3T3-L1 cells. Two hundred microliter of pre-adipocyte medium (PM-1) containing 3,000 cells was dispensed into each well of a 96-well, tissue culture plates. The assay plates were incubated at 37 °C, 95% humidity and 5% CO2 for 3 days. The differentiation of pre-adipocytes to adipocytes was induced by removing the pre-adipocyte medium and replacing it with 200 μL of differentiation medium (DM-2). One μL of test compounds at different concentrations dissolved in differentiation medium were added to the assay plates. The final DMSO concentration was 0.5%. Triacsin C was used as a positive control for lipid under-storage. After 3 days, the differentiation medium was aspirated and replaced with 200 μL of adipocyte maintenance medium (AM-1), and the assay plates were incubated for additional 4 days. Cells were then fixed with 4% paraformaldehyde in PBS for 30 minutes at room temperature, washed 3 times with PBS buffer and stained with 200 μL of PBS containing BODIPY lipid dye 493/530, Hoechst 33342 and CellMask deep red dye. Images were acquired using the GE InCell 2000 imager with 20× magnification. The number of cells, number of lipids, and size of lipids were quantitated and determined using the GE Investigator software.
2.2. Probe Chemical Characterization

Probe ML360 (CID 70789742)
*Purity > 95% as determined by LC/MS and 1H NMR analyses.
N-(3,4-diethoxyphenethyl)-1-(3-methylbenzyl)-1H-pyrazole-5-carboxamide:1H NMR (400 MHz, DMSO-d6) δ 8.53 (t, J = 5.7 Hz, 1H), 7.51 (d, J = 2.0 Hz, 1H), 7.16 (t, J = 7.6 Hz, 1H), 7.05 (dddd, J = 7.6, 1.9, 1.2, 0.7 Hz, 1H), 6.96 (ddq, J = 1.8, 1.3, 0.6 Hz, 1H), 6.89 (dtt, J = 7.6, 1.3, 0.7 Hz, 1H), 6.83 (s, 1H), 6.81 (d, J = 6.5 Hz, 1H), 6.77 (d, J = 2.0 Hz, 1H), 6.66 (dd, J = 8.1, 2.0 Hz, 1H), 5.68 (s, 2H), 4.00 – 3.89 (m, 4H), 3.39 (dt, J = 7.8, 6.2 Hz, 2H), 2.71 (t, J = 7.3 Hz, 2H), 2.24 (d, J = 0.7 Hz, 3H), 1.28 (dt, J = 7.6, 7.0 Hz, 6H). Method 1, retention time: 6.183 min; HRMS: m/z (M+H)+ = 408.2269 (Calculated for C24H30N3O3 = 408.2282).

Figure 1Stability of ML360 in different assay buffers in > 50 hr time course
(A) Stability of 200 μM ML360 in PBS buffer pH 7.4 showed slight decrease in signal; however, the loss of UV signal was determined to be due to precipitation of the probe because of the high concentration needed for the assay. (B) Stability of ML360 in aqueous PBS buffer containing 5 mM glutathione and (C) in the medium used for the 3T3-L1 cell culture experiments showed no significant changes over time.
Figure 2Structures of ML360 and analogs with their corresponding Compound IDs listed in Table 1
Table 1
List of probe ML360 and related analogs.
2.3. Probe Preparation
Preparation of N-(3,4-diethoxyphenethyl)-1-(3-methylbenzyl)-1H-pyrazole-5-carboxamide (ML360) is a three-step process described below and illustrated in Scheme 1.

Scheme 1
Synthetic route to ML360 is a 3 step process. The crude product of the last step is further purified by reverse phase chromatography to give the ML360 probe and its regioisomer as TFA salts.
- Potassium carbonate (K2CO3) and 1-(bromomethyl)-3-methylbenzene were added to a solution of methyl 1H-pyrazole-3-carboxylate in dimethylformamide (DMF). The mixture was stirred at room temperature (r.t.) overnight. Water was added to the mixture and extracted with ethyl acetate (EtOAc) twice. The organic layer was washed with saturated sodium bicarbonate (NaHCO3) and brine, dried over sodium sulfate (Na2SO4) and concentrated to give the crude product used in the next reaction without purification.
- Lithium hydroxide (LiOH) was added to a solution of methyl 1-(3-methylbenzyl)-1H-pyrazole-3-carboxylate and methyl 1-(3-methylbenzyl)-1H-pyrazole-5-carboxylate in tetrahydrofuran (THF). The mixture was stirred at r.t. overnight. The solvent was removed giving a crude product that was used in the next reaction without purification.
- 2-(3,4-diethoxyphenyl)ethanamine was added to a solution of 1-(3-methylbenzyl)-1H-pyrazole-3-carboxylic acid, 1-(3-methylbenzyl)-1H-pyrazole-5-carboxylic acid, O-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HATU) and N,N-Diisopropylethylamine (DIPEA) in DMF. The mixture was stirred at r.t. for 2 hours. Water was added to the mixture, and extracted with EtOAc twice. The organic layer was washed with saturated NaHCO3 and brine, dried over Na2SO4, filtered and concentrated. The crude product was purified by reverse phase chromatography to give ML360 and its regioisomer as TFA salts.
3. Results
3.1. Dose Response Curves for Probe

Figure 3Dose response curves for the probe ML360 (green) and negative control compound 1 (blue)
Results showed dose response reduction of lipid drop formation in Drosophila S3 Cells S3 (AID 652175) upon treatment with ML360 but not with the compound 1.
3.2. Cellular Activity

Figure 4Additional cellular activity evaluation of the probe ML360
(A) Lipid droplet accumulation in mouse 3T3-L1 cells (AID 652180) showed dose response inhibition of ML360 (green circle) but not with the negative control compound 1 (blue circle). (B) 3T3-L1 differentiation assay (AID 652183) showed dose response reduction of the lipid droplets (blue square) without any cytotoxic effect as the cell number per treatment remained the same (orange circle).
3.3. Profiling Assays
Preliminary absorption, distribution, metabolism, and excretion (ADME) profiling of ML360 demonstrated good stability in glutathione and PBS buffers for > 48 hr. Results also showed reasonable permeability and liver microsome stability (Table 2).
Table 2
In vitro ADME profiling of ML360.
4. Discussion
4.1. Comparison to Existing Art and How the New Probe is an Improvement
The control for the original HTS and subsequent assays was Triacsin C (Figure 5), a previously reported inhibitor of LDs [6]. Triacsin C is reported to be an inhibitor of long-chain fatty acyl CoA synthetase and based on its structure, likely inhibit additional lipid synthetases. Three unique chemotypes (Figure 5) previously described are also observed to be capable of potently reducing the LD phenotype, including a tetrahydro-1H-indazole series (ML206), a diethoxyphenethyl-thienopyrrole series (ML219) and a ureido-piperidine-4-carboxylate ester series (ML220) [7]. All analogs showed sub-micromolar potency in reducing LDs in Drosophila melanogaster S3 cells. However unlike the old probes, ML360 retained potent activity in 3T3-L1 pre-adipocyte differentiation conditions and other mammalian cell lines without toxic effect.
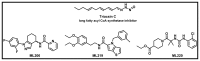
Figure 5
Previously reported probes that inhibit lipid droplet formation.
5. References
- 1.
- Kühnlein R. Thematic review series: Lipid droplet synthesis and metabolism: from yeast to man. Lipid droplet-based storage fat metabolism in Drosophila. Journal of Lipid Research. 2012;53(8):1430–1436. [PMC free article: PMC3540836] [PubMed: 22566574]
- 2.
- Farese R, Walther T. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139(5):855–860. [PMC free article: PMC3097139] [PubMed: 19945371]
- 3.
- Mak H. Lipid droplets as fat storage organelles in Caenorhabditis elegans: Thematic Review Series: Lipid Droplet Synthesis and Metabolism: from Yeast to Man. Journal of Lipid Research. 2012;53(1):28–33. [PMC free article: PMC3243478] [PubMed: 22049244]
- 4.
- Zechner R, et al. FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell Metabolism. 2012;15(3):279–291. [PMC free article: PMC3314979] [PubMed: 22405066]
- 5.
- Winchester B, Vellodi A, Young E. The molecular basis of lysosomal storage diseases and their treatment. Biochemical Society Transactions. 2000;28(2):150–154. [PubMed: 10816117]
- 6.
- Igal R, et al. Triacsin C blocks de novo synthesis of glycerolipids and cholesterol esters but not recycling of fatty acid into phospholipid: evidence for functionally separate pools of acyl-CoA. The Biochemical Journal. 1997;324(2):529–534. [PMC free article: PMC1218462] [PubMed: 9182714]
- 7.
- Boxer M, et al. Modulators of Lipid Storage Probe Reports from the NIH Molecular Libraries Program. http://www
.ncbi.nlm.nih .gov/books/NBK47352/
- PMCPubMed Central citations
- PubChem BioAssay for Chemical ProbePubChem BioAssay records reporting screening data for the development of the chemical probe(s) described in this book chapter
- PubChem SubstanceRelated PubChem Substances
- PubMedLinks to PubMed
- Review Modulators of Lipid Storage.[Probe Reports from the NIH Mol...]Review Modulators of Lipid Storage.Boxer MB, Shen M, Zhang Y, Liu L, Auld DS, Beller M. Probe Reports from the NIH Molecular Libraries Program. 2010
- Models of lipid droplets growth and fission in adipocyte cells.[Exp Cell Res. 2015]Models of lipid droplets growth and fission in adipocyte cells.Boschi F, Rizzatti V, Zamboni M, Sbarbati A. Exp Cell Res. 2015 Aug 15; 336(2):253-62. Epub 2015 Jun 26.
- Simulating the dynamics of lipid droplets in adipocyte differentiation.[Comput Methods Programs Biomed...]Simulating the dynamics of lipid droplets in adipocyte differentiation.Boschi F, Rizzatti V, Zamboni M, Sbarbati A. Comput Methods Programs Biomed. 2017 Jan; 138:65-71. Epub 2016 Oct 26.
- Lipid droplets fusion in adipocyte differentiated 3T3-L1 cells: a Monte Carlo simulation.[Exp Cell Res. 2014]Lipid droplets fusion in adipocyte differentiated 3T3-L1 cells: a Monte Carlo simulation.Boschi F, Rizzatti V, Zamboni M, Sbarbati A. Exp Cell Res. 2014 Feb 15; 321(2):201-8. Epub 2014 Jan 3.
- Review The size matters: regulation of lipid storage by lipid droplet dynamics.[Sci China Life Sci. 2017]Review The size matters: regulation of lipid storage by lipid droplet dynamics.Yu J, Li P. Sci China Life Sci. 2017 Jan; 60(1):46-56. Epub 2016 Dec 13.
- ML360, A Potent Inhibitor of Lipid Droplet Formation - Probe Reports from the NI...ML360, A Potent Inhibitor of Lipid Droplet Formation - Probe Reports from the NIH Molecular Libraries Program
Your browsing activity is empty.
Activity recording is turned off.
See more...

