Downloading dbGaP data with JWT or NGC
Introduction
The database of Genotypes and Phenotypes (dbGaP) was developed to archive and distribute the data and results from studies investigating the interaction of genotype and phenotype in humans.
The following guide outlines how you configure the SRA Toolkit for accessing protected data from dbGaP. Detailed information regarding the usage of individual tools in the SRA Toolkit can be found on the tool-specific documentation pages.
Prerequisites
- Users must have the latest version of the SRA Toolkit installed.
- Users that wish to access controlled-access data must first apply for approval. Please review the process at the Authorized Access Portal
 .
. - Once granted access to a project, the PI may login to the Run Selector and access specific dbGaP study or studies using the phs identifier.
Getting the JWT Cart and using it in the Amazon Web Services (AWS) and Google Cloud Platform (GCP)
Obtain jwt.cart file
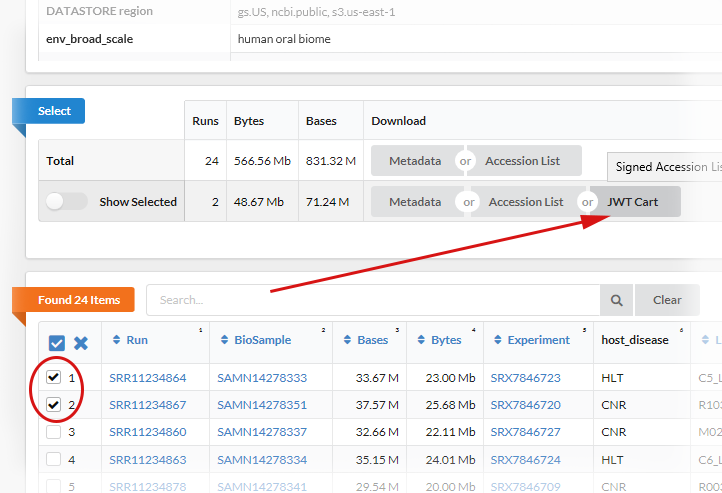
In the Run Selector select at least one Run to activate the button JWT Cart.
Press the button to download the JWT Cart file, please be aware that the file has an 1 hour expiration time.
Move the jwt.cart file to the Virtual Machine (VM) instance by using text editor Nano
The easiest way of moving the JWT file to your VM instance is to open the file in a text editor and copy and paste the content to the Nano Text Editor.
- Create the file in the VM:
nano jwt.cart - Copy the content of the jwt.cart (
Ctrl+AandCtrl+C) and paste it by using the right mouse button into Nano - Press
Ctrl+S(to save the file) and then,Ctrl+Xto exit.
Download SRA data
Once the JWT file is in your instance, you can download all the accessions using the prefetch utility:
prefetch --perm jwt.cart
You can also selectively download the data with this command:
./prefetch --perm jwt.cart SRR1219879
Alternatively, you can download/convert the data into your format of choice (SAM, FASTQ, etc.) using the dump utilities:
./fasterq-dump --perm jwt.cart SRR1219879
Downloading with NGC for use on any server
Starting with SRA Toolkit version 2.10.2, there are several important changes:
- You no longer need to import the NGC file to the configuration
- The NGC file will need to be specified as part of the command line every time you run a tool
- For SRA Runs, you no longer have an option to create a cart, but will need to use a list of Run accessions
Engage
NCBI wants your feedback on SRA in the Cloud. Contact sra@ncbi.nlm.nih.gov with questions or if you would like to provide input on new functionality.
