NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
At least six cellular pathways leading to activation of NFκB-family transcription factors have been identified, participating in host-defense, immunity, inflammation, and cancer. The pathway initiated by Protein Kinase C (PKC) family kinase is critical for signaling downstream of antigen receptors in T-cells and B-cells, as well as many growth factor receptors and some GPCRs that stimulate intracellular Ca2+ elevations. This pathway often involves a multi-component protein complex that includes CARMA, Bcl-10, and MALT1. Previously, a potent (~67 nM) and selective probe (ML029, Pubchem CID2858522) was identified as an inhibitor of the PKC-initiated NF-κB pathway after a battery of 12 secondary and counter-screen assays. However, this probe appeared potent only in the original HEK293 reporter cell line and some types of primary lymphocytes, with little potency in several other tumor cell lines. We used HTS assays employing NF-κB reporter genes in T-cell and B-cell lines for screening the ~330,600 compound MLSMR collection to identify novel pathway-selective inhibitors. We now disclose the discovery and characterization of two novel potent small molecule cell-active NF-κB inhibitors that are both (a) selective for the NF-κB pathway induced by PKC activators (phorbol myristic acetate/ionomycin) and (b) selective for B-cells. The first oxadiazole-based probe (ML236, PubChem CID665654) is potent (35 nM) in a B-cell line and >400-fold selective over both the NF-κB activation in T-cells and against TNFα-mediated NF-κB activation. In comparison, the second oxazole-based probe (ML237, PubChem CID4174238) is also potent (208 nM) in a B-cell line and >400 fold selective over both the NF-κB activation in T-cells and against TNFα-mediated NF-κB activation. The synthetic methodology, SAR development, and initial pharmacologic characterization are also presented. These probe molecules will be used to define the B-cell specific targets along these pathways and will serve as tools to reveal hitherto unknown components of these signaling pathways. These probes will prove useful to researchers working in immunology, autoimmune diseases, transplantation and allograft rejection, lymphoid malignancies, immune signaling, and general signal transduction.
Assigned Assay Grant #: 1 X01 MH077633-01
Screening Center Name & PI: Sanford Burnham Center for Chemical Genomics (SBCCG) & John C. Reed (PI)
Chemistry Center Name & PI: same as above
Assay Submitter & Institution: John C. Reed, Sanford-Burnham Medical Research Institute
PubChem Summary Bioassay Identifier (AID): 435012
Probe Structure & Characteristics
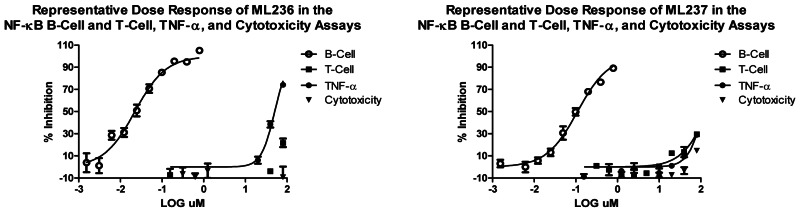
This Center Probe Report describes two novel inhibitors of NF-κB activation in B-cells, ML236 (CID665654) and ML237 (CID4174238). Potency, selectivity and cytotoxicity characteristics are summarized for these probes below.
| CID/ML# | Target Name | IC50 (nM) [SID, AID] | Anti-target Name(s) | IC50 (μM) (selectivity) [SID, AID] |
|---|---|---|---|---|
| CID665654 ML236 | NF-κB activation in B-cells | 34.9 nM SID103061970 AID504485, 504487, 504489 | NF-κB activation in T-cells | >79 μM (>2263X) SID103061970 AID504512, 504514, 504516 |
| Oxadiazole | TNFα-mediated NF-κB activation | 53.2 μM (>1524X) SID103061970 AID504506, 504517, 504518 | ||
| Cytotoxicity | >79 μM SID103061970 AID493251 | |||
| CID4174238 ML237 | NF-κB activation in B-cells | 190 nM SID103061890 AID504485, 504487, 504489 | NF-κB activation in T-cells | >79 μM (>379X) SID103061890 AID504512, 504514, 504516 |
| oxazole | TNFα-mediated NF-κB activation | >79 μM (>379X) SID103061890 AID504506, 504517, 504518 | ||
| Cytotoxicity | >79 μM SID103061890 AID493251 | |||
Recommendations for scientific use of the probe
Regulated control of NF-κB is critical for the immune, inflammatory, proliferative, anti-apoptotic, and differentiation responses in both T- and B-cells via the ligation of their respective antigen receptors in confluence with a number of other cell specific co-receptor activities. The signaling that leads to the various cell mediated outcomes is complex and the mechanisms by which NF-κB activation performs specific functions that drive these outcomes is not thoroughly understood. Although many biological tools exist for probing NF-κB functions, there are few if any small molecule tools that can probe the common non-canonical TCR and BCR signaling pathways leading to NF-κB activation. This NF-κB pathway activated downstream of antigen receptors is uniquely required for adaptive immune response, setting it apart from the numerous NF-κB pathways implicated in innate immunity. Probes that are discovered by the screening for compounds that inhibit TCR and BCR mediated NF-κB activation will be used to define the specific targets along these pathways and will serve as tools to reveal hitherto unknown components of these pathways. These probes will prove useful to researchers working in immunology, autoimmune diseases, transplantation and allograft rejection, lymphoid malignancies, immune signaling, and general signal transduction.
1. Introduction
Overall Goal of Project
This assay was originally assigned to the Scripps Research Institute Molecular Screening Center (TSRI MSC) by the NIH during the MLSCN pilot phase. The Scripps Center completed primary screening (AID: 465) on only 61,609 compounds of the NIH compound collection, as well as dose-response experiments (PubChem AID: 586). Unfortunately, none of the compounds identified from the gene reporter assay suppressed the secretion of IL-8, an endogenous NF-κB inducible gene product in secondary assays in relevant cell lines. Dr. Reed’s lab screened an additional 53,280 from his internal collection and was ultimately able to identify one pathway selective probe molecule (CID2858522-ML029), after a battery of 12 secondary and counter-screen assays. The rationale, assays, flowchart for hit validation, and SAR support to the eventual probes are detailed in the Chemical Probe Report (CPR) that can be accessed from the MLP site: http://mli.nih.gov/mli/mlp-probes/ or the full pdf can be downloaded from the following URL: https://mli.nih.gov/mli/?dl_id=454. This prior probe, ML029, appears to only be potent in the original HEK293 reporter cell line and shows poor or little potency in HEK293T (containing the SV40 T antigen) or other more relevant cell lines.
As summarized in the CPR for ML029, the previous probe compound inhibits the activity of the NF-κB luciferase reporter gene in HEK-293 cells stimulated with PMA and Ionomycin with IC50 < 100 nM. The compound is similarly active using phorbol dibutryate (PDB) instead of PMA (phorbol myristic acetate) as a PKC agonist. At concentrations up to 4 μM, the compound is inactive at suppressing NF-κB luciferase reporter gene activity induced in the same cells (HEK293) by TNFα or by transfection with an active version of TLR4 (CD4-TLR4 fusion protein; member of Toll-like Receptor family) or by transfection with CD40 (a TNF-family receptor), or by transfection with NOD1, or by transfection with NOD2, or by transfection of PIDD (p53-induced protein with a death domain), a protein playing a critical role in DNA-damage induced NF-κB activation. The probe compound also inhibits PMA-induced secretion of the NF-κB inducible cytokine IL-8 by HEK293 cells, with IC50 ~ 100 nM. The compound is not toxic against HEK293, HEK293T, THP.1, Jurkat, MCF7, or HeLa cells. In contrast to HEK293 cells, the probe compound is inactive with respect to suppression of PMA-induced NF-κB reporter gene activity in HEK293T cells, HeLa, MCF7, and PPC-1 cells. The probe compound partially inhibits IL-2 production by Jurkat T-cells stimulated with anti-CD3/antiCD28, consistent with partial dependence on NF-κB for T-cell antigen receptor-mediated induction of the IL-2 gene.
The NIH collection has grown to >325,000 compounds since the original MLSCN screen of 61,609 of the MLSMR. The goal therefore is to identify additional compounds that selectively inhibit one of the several known pathways that lead to NF-κB activation in relevant mammalian cells in a T-cell or B-cell specific manner. In the original MLSCN screen a stably transfected reporter epithelial cell line (HEK293) that contains a luciferase gene driven by a NF-κB responsive promoter was used for compound library screening. The hits identified from that screen were very much less potent when tested in B and T cell lines. Therefore, in this rescreening paradigm, the primary screen(s) will be accomplished with two separate but related screens using an NF-κB-luciferase reporter cloned into a T-cell line (Jurkat) or a B-cell line (697). The data from these two separate screens were used to define three classes of potent (<1 μM IC50) cell-type specific small molecule chemical inhibitors of NF-κB activation: (1) T-cell specific, (2) B-cell specific, and (3) T & B-cell dual specific. These cell-type selective inhibitors are however still to be selective against TNF (>10 X potency), inhibit IL-8 secretion at comparable potencies, and are selective (>10X potency) against representative PKCs (PKC beta and PKC theta that are involved in T and B cell signaling).
As before, at least six cellular pathways leading to activation of NF-κB-family transcription factors have been identified, participating in host-defense, immunity, inflammation, and cancer. A pathway activated by antigen receptors on T- and B-lymphocytes has been revealed, involving a cascade of participating proteins that includes CARMA1 (BIMP3), Bcl-10, paracaspase (MALT1), TRAF6, and Ubc13. This pathway is initiated by Protein Kinase C-theta, which induces phosphorylation of components of this signaling pathway (reviewed in Thome 2004). Based on experiments using siRNA and dominant negative mutants, it was determined that treatment of cells with the combination of phorbol ester Phorbol Myristic Acetate (PMA) and Calcium-ionophore Ionomycin triggers this pathway, resulting in NF-κB activation (Ruland et al. 2001; McAllister-Lucas et al. 2001; Rueffli-Brasse et al. 2003; Zhou et al. 2004). Counter-screens using activators of other NF-κB activation pathways identify pathway specific inhibitors, including (1) NF-κB reporter gene activity or cytokine production induced by Lipopolysaccharide (LPS) in the THP.1 monocyte cell line (TLR4 agonist), (2) by gamma-Tri-DAP and muramyl dipeptide (MDP) in MCF7 breast cancer or HCT116 colon cancer cells (NLR agonists), (3) DNA damaging agent Doxorubicin in Jurkat and 697 cells, and (4) BAFF (a TNF family member that induces a non-canonical, alternative NF-κB pathway) in 697 cells.
In summary, Dr. Reed’s lab seeks potent (<1 μM IC50) chemical inhibitors of NF-κB activation that are (1) T-cell specific, (2) B-cell specific, or (3) T & B-cell dual specific, that are selective against TNF and their cognate PKCs (>10 X), and inhibit IL-8 secretion at comparable potencies.
Aims of the project: We propose to use this cell-based HTS assay for screening the NIH’s chemical library. The compounds identified using this assay will be useful research tools for analysis of the physiological roles of this NF-κB activation pathway.
2. Materials and Methods
The details of the primary HTS and additional assays can be found in the “Assay Description” section in the PubChem BioAssay view under the AIDs as listed in Table 1 below. Additionally the details for the primary HTS are provided in the Appendix at the end of this probe report.
Table 1
Summary of Assays and AIDs.
2.1. Assays
Table 1 summarizes the details for the assays that drove this probe project.
2.2. Probe Chemical Characterization
a. Chemical name of probe compound
The IUPAC names of the probes are 3-(3-(4-(tert-butyl)phenyl)-1,2,4-oxadiazol-5-yl)-N-methylpropanamide (ML236) and N-cyclopropyl-2-(((2-(4-ethoxyphenyl)-5-methyloxazol-4-yl)methyl)thio)acetamide (ML237).
The specific batches prepared, tested and submitted to the MLSMR are archived as SID103061876 corresponding to CID665654 (ML236) and SID103061890 corresponding to CID4174238 (ML237).
b. Probe chemical structure including stereochemistry if known
c. Synthesis and Structural Verification Information of probe SID103061876 corresponding to CID665654 (ML236)

Scheme 1Synthesis of ML236, conditions
a. Hydroxylamine hydrochloride, triethylamine, methanol, 80 °C, 30 minutes; b. succinic anhydride, DMF, 120 °C, overnight, 47% for two steps; c. thionyl chloride, methylene chloride, 0 °C to room temperature, 3 hours; d. methylamine, methylene chloride, 0 °C to room temperature, overnight, 85% for two steps.
Synthesis and Structural Verification Information of probe SID103061890 corresponding to CID4174238 (ML237)
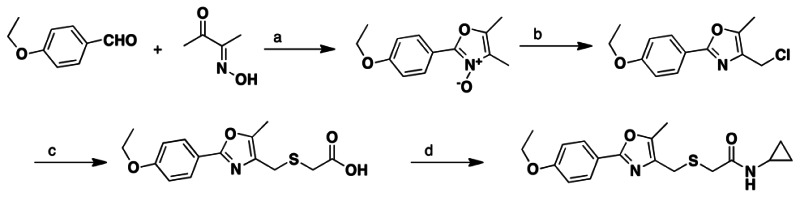
Scheme 2Synthesis of ML237, conditions
a. acetic acid, hydrochloric acid (aq), 0 °C, 3 hours, 23%; b. POCl3, chloroform, refluxing, 2 hours, 80%; c. thioglycolic acid, sodium hydroxide, methanol, 65 °C, 2 hours, 79%; d. cyclopropylamine, EDCI, HOBt, diisopropylethylamine, methylene chloride, 0 °C to room temperature, overnight, 80%.
For detailed procedures see Section 2.3. Images of spectral data (1HNMR, 13CNMR, and LC/MS) used to support the structural assignment of ML236 and ML237 can be found in Section 6 (Supplementary Information).
d. If available from a vendor, please provide details
Samples of ML236 and ML237 synthesized at SBCCG have been deposited in the MLSMR (Bio-Focus DPI) (see Probe Submission Tables 3a/b below). ML236 is available from Ambinter, Cat#: STK86117, ambinter.com; ML237 is available from Aurora, Cat# K01.550.497, aurorafinechemicals.com.
Table 3a
Probe and Analog Submissions to MLSMR (BioFocus DPI) for B-cell Specific Inhibitors of NF-κBActivation.
Table 3b
Probe and Analog Submissions to MLSMR (BioFocus DPI) for B-cell Specific Inhibitors of NF-κBActivation.
e. Solubility and Stability of probe in PBS at room temperature
The stability and solubility of ML236 and ML237 was investigated in PBS buffer at room temperature (Figure 1). ML236 showed slight instability after 48 hr. in PBS (80% remaining), whereas ML237 was completely stable (see Figures and Table below)

Figure 1
Stability of ML236 and ML237 in PBS.
f. A tabulation of calculated and known probe properties
Table 2Calculated Probe Properties for ML236 & ML237
| Calculated Property | Value | |
|---|---|---|
| ML236 CID665654 | ML237 CID4174238 | |
| Molecular Weight [g/mol] | 287.3568 | 346.4439 |
| Molecular Formula | C16H21N3O2 | C18H22N2O3S |
| XLogP3-AA | 3 | 3.1 |
| H-Bond Donor | 1 | 1 |
| H-Bond Acceptor | 4 | 4 |
| Rotatable Bond Count | 5 | 8 |
| Tautomer Count | 6 | 2 |
| Exact Mass | 287.1634 | 346.1351 |
| MonoIsotopic Mass | 287.1634 | 346.1351 |
| Topological Polar Surface Area | 68 | 89.7 |
| Heavy Atom Count | 21 | 24 |
| Formal Charge | 0 | 0 |
| Complexity | 346 | 411 |
| Isotope Atom Count | 0 | 0 |
| Defined Atom StereoCenter Count | 0 | 0 |
| Undefined Atom StereoCenter Count | 0 | 0 |
| Defined Bond StereoCenter Count | 0 | 0 |
| Undefined Bond StereoCenter Count | 0 | 0 |
| Covalently-Bonded Unit Count | 1 | 1 |
Samples of the probes (>25 mg) and each of five analogs (>20 mg) synthesized at SBCCG were submitted to the MLSMR (Tables 3a and 3b), and 5 mg of the probe was provided to Dr. Reed’s lab.
2.3. Probe Preparation
ML236 Preparation
Step 1. A microwave vessel was charged with 4-(tert-butyl)benzonitrile (1.06 g, 6.66 mmol) and methanol (10 mL). Triethylamine (1.03 mL, 6.66 mmol) and hydroxylamine hydrochloride (460.0 mg, 6.66 mmol) were added. The tube was put in Biotage microwave reactor and was heated at 80 °C for 30 minutes. The mixture was transferred to a round bottom flask and the solvent was removed under reduced pressure. The flask was put under high vacuum for 30 minutes. The solid was dissolved in N, N′-dimethylformamide (10 mL). Succinic anhydride (880.0 mg, 6.66 mmol) was added. The mixture was stirred at 120 °C overnight. The solution was cooled down to room temperature. Hydrochloride solution (60 mL, 1N solution) was added. The mixture was extracted with ethyl acetate (3 × 40 mL), washed with brine (60 mL), dried over anhydrous sodium sulfate, filtered and concentrated under reduced pressure. The crude product was titrated with hexane (100 mL) and filtered. The white solid (850.0 mg, 47% for two steps) was used in the next step without further purification. 1H NMR (400 MHz, CDCl3) δ 7.97 (d, J = 10.5 Hz, 2H), 7.49 (d, J = 10.3 Hz, 2H), 3.28 (t, J = 7.4 Hz, 2H), 3.05 (t, J = 7.4 Hz, 2H), 1.35 (s, 3H). ESI m/z 275 [M+H].
Step 2. A round-bottom flask was charged with 3-(3-(4-(tert-butyl)phenyl)-1,2,4-oxadiazol-5-yl)propanoic acid (314.0 mg, 1.24 mmol) and methylene chloride (10.0 mL) at 0 °C under N2 atmosphere. Thionyl chloride (0.18 mL, 2.48 mmol, 2.0 equiv.) was added dropwise. The resulting solution was stirred for 30 minutes at 0 °C. The mixture was then stirred at room temperature for 3 hour. The solvent was removed under reduced pressure and the flask was put under high vacuum for 30 minutes. The solid was dissolved in methylene chloride (10 mL) at 0 °C under N2 atmosphere. Methylamine solution (2M solution in tetrahydrofuran, 1.3 mL, 2.48 mmol, 2.0 equiv.) was added dropwise. The mixture was stirred for 1 hour at 0 °C. The solution was diluted with methylene chloride (30 mL) and was transferred to a separation funnel. The mixture was washed with saturated ammonia chloride solution (30 mL). The organic and aqueous layers were separated. The aqueous layer was extracted with methylene chloride (2 × 40 mL). The combined organic layer was washed with brine (30 mL), dried over anhydrous sodium sulfate, filtered and concentrated under reduced pressure. The crude product was purified by column chromatography eluting with 30–50% ethyl acetate/hexane to afford the desired product as a white solid (302.9 mg, 85% yield): 1H NMR (400 MHz, CDCl3) δ 7.98 (d, J = 10.5 Hz, 2H), 7.53 (d, J = 10.3 Hz, 2H), 5.77 (br, s, 1H), 3.32 (t, J = 7.4 Hz, 2H), 2.86 (d, J = 4.9 Hz, 3H), 2.79 (t, J = 7.4 Hz, 2H), 1.37 (s, 3H). 13C (100 MHz, CDCl3) δ 178.7, 170.9, 168.2, 154.6, 127.2, 125.8, 123.8, 34.9, 32.3, 31.2, 26.5, 22.5. Melting point: 120–122 °C.
ML237 Preparation
Step 1. A solution of 2,3-butanedione monooxime (5.80 g, 58.3 mmol) and 4-ethoxybenzaldehyde (9.30g, 62.0 mmol) in acetic acid (50 mL) was cooled down to 0 °C and HCl solution (24 mL, 96.0 mmol, 4 M in 1,4 dioxane solution) was added. The mixture was stirred at 0 °C for 3 hour. Diethyl ether (100 mL) was added to the mixture and the resulting slurry was stirred for 1 hour at 0 °C. The white precipitate was filtered and washed with ether (20 mL). The solid was transferred to a beaker and washed with water (100ml) and concentrated ammonia hydroxide solution (12 mL). The mixture was extracted with CHCl3 (2 × 100 mL), washed with brine (100 mL), dried over anhydrous sodium sulfate, filtered and concentrated under reduced pressure to afford 2-(4-ethoxyphenyl)-4,5-dimethyloxazole-3-oxide as a white solid (2.8 g, 23% yield). The unstable compound was used in the next step without further purification.
Step 2. To a solution of 2-(4-ethoxyphenyl)-4,5-dimethyloxazole-3-oxide (2.8g, 12.0 mmol) in anhydrous CHCl3 (20 ML) was added POCl3 (1.2 mL, 13.2 mmol) dropwise at room temperature. The mixture was heated to reflux for 2 hour, cooled to room temperature, poured into ice water (100 mL), neutralized with saturated NaHCO3 solution (100 mL). The mixture was extracted with CHCl3 (2 × 100 mL), washed with brine and dried over anhydrous sodium sulfate, filtered and concentrated under reduced pressure. The crude product was dissolved in hot hexane (100 mL), decanted from the insoluble dark material and cooled to 0 °C to allow the product to crystallize out. The solid was filtered and dried. Yield: 2.40 g (80%). 1H NMR (400 MHz, CDCl3) δ 7.95 (d, J = 6.9 Hz, 2H), 6.95 (d, J = 8.9 Hz, 2H), 4.11 (q, J = 6.9 Hz, 2H), 3.81 (s, 3H), 3.38 (s, 2H), 2.39 (s, 3H), 1.48 (t, J = 6.9 Hz, 3H). ESI m/z 252 [M+H].
Step 3. A solution of NaOH (770.0 mg, 19.2 mmol) and thioglycolic acid (883.0 mg, 9.6 mmol) in MeOH (30 mL) was stirred at room temperature for 30 minutes. 4-(chloromethyl)-2-(4-ethoxyphenyl)-5-methyloxazole (2.40 g, 9.6 mmol) was added and the mixture was stirred at 65 °C for 2 hour, cooled to room temperature. The solvent was removed under reduced pressure. Hydrochloride solution (100 mL, 1N solution) was added. The mixture was extracted with ethyl acetate (2 × 80 mL), washed with brine and dried over anhydrous sodium sulfate, filtered and concentrated under reduced pressure. The crude product was titrated with hexane (100 mL). The solid was filtered and dried. Yield: 2.3 g (79%). 1H NMR (400 MHz, CDCl3) δ 7.92 (d, J = 6.9 Hz, 2H), 6.92 (d, J = 8.9 Hz, 2H), ESI m/z 308 [M+H].
Step 4. A round-bottom flask was charged with 2-(((2-(4-ethoxyphenyl)-5-methyloxazol-4-yl)methyl)thio)acetic acid (270.0 mg, 0.88 mmol) and methylene chloride (6.0 mL) at 0 °C. N,N-Diisopropylethylamine (0.31 mL, 1.76 mmol, 2.0 equiv.) was added followed by addition of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (260.0 mg, 1.32 mmol, 1.50 equiv.) and hydroxybenzotriazole (180.0 mg, 1.32 mmol, 1.50 equiv.). The resulting solution was stirred for 30 minutes at 0 °C. Cyclopropylamine (0.12 mL, 1.76 mmol, 2.0 equiv.) was added and the solution was stirred at room temperature overnight. The solution was diluted with methylene chloride (30 mL) and was transferred to a separation funnel. The mixture was washed with saturated ammonia chloride solution (30 mL). The organic and aqueous layers were separated. The aqueous layer was extracted with methylene chloride (2 × 30 mL). The organic layer was combined and washed with brine (30 mL). The organic layer was dried over anhydrous sodium sulfate, filtered and concentrated under reduced pressure. The crude product was purified by column chromatography eluting with 30–50% ethyl acetate/hexane to afford the desired product as a white solid (243.4 mg, 80% yield): 1H NMR (400 MHz, CDCl3) δ 7.89 (d, J = 6.9 Hz, 2H), 7.45 (br s, 1H), 6.95 (d, J = 8.9 Hz, 2H), 4.09 (q, J = 6.9 Hz, 2H), 3.58 (s, 2H), 3.13 (s, 2H), 2.63 (m, 1H), 2.33 (s, 3H), 1.42 (t, J = 6.9 Hz, 3H), 0.71 (m, 2H), 0.43 (m, 2H). 13C (100 MHz, CDCl3) δ 170.6, 160.9, 160.4, 145.1, 132.1, 127.8, 120.1, 114.8, 63.8, 35.4, 27.6, 22.9, 14.9, 10.3, 6.5. Melting point: 125–126 °C.
3. Results
3.1. Summary of Screening Results
The flowchart on the right summarizes (Figure 2) the screening results for the NF-κB B- and T-Cell Assays. Following primary HTS of approximately 330,000 Molecular Libraries Small Molecules Repository (MLSMR) compounds at 4 μM in the NF-κB B-Cell Assay (AID435022), 1085 initial actives (~0.33% hit rate) were obtained at a 35% activity cut-off. For a cell-based assay, the screen was robust with an average Z′ of 0.68.
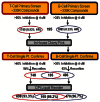
Figure 2
Screening results and Hit validation.
The 1085 compound hits from the screen were cherry-picked in-house and retested in the NF-κB B-Cell assay confirmatory assay (AID449746) at 4 μM. For the single point re-confirmatory assay, a cut-off of 30% inhibition yielded a 63% reconfirm rate resulting in a total of 486 compounds. Fresh stock solution “cherry picks” of the 486 hits were requested from the MLSMR and 470 were received (96.7%) and tested and activities (IC50) were confirmed in full 10-point dose-response titrations (AID489023). In parallel, selectivity was determined against the Jurkat (T-Cell) (AID489006) cell line and against the TNFα modulated pathway (AID489022). Also, the cytotoxicity of the compound hits was evaluated (AID489021). Based on the experimental results, dry powder compounds were selected for follow up studies and additional, commercially available analogues were ordered. From this we identified two series of compounds that were B-cell specific inhibitors of NF-κB activation: (1) an oxadiazole, CID665654 and (2) an oxazole, CID4174238, that both had <0.5 μM potency for inhibition of NF-κB activation in B-cells with > 80 μM potency for NF-κB activation in T-cells, and > 80 μM potency for TNFα mediated NF-κB activation.
3.3. Scaffold/Moiety Chemical Liabilities
Neither the oxadiazole and oxazole scaffolds contain no standard chemical liabilities, nor do the lead molecules appear to be frequent hitters. Some drug discovery efforts around similar scaffolds have been reported (6).
3.4. SAR Tables
The general SAR strategy we pursued around the oxadiazole scaffold from the screening hit, CID665654 (entry 7 in Table 4) is depicted in Figure 4. Efforts were made to expand the SAR around both the aryl group (in green) as well to investigate the SAR of the amide moiety, including examining various esters as well as the parent acid (in blue). The results are covered in Tables 4 and 5 below.
Table 4
SAR Analysis of B-cell Specific Inhibitors of NF-κB Activation: Oxadiazoles.

Figure 4
General SAR strategy around oxadiazole scaffold.
Table 5
SAR Analysis of B-cell Specific Inhibitors of NF-κB Activation: Oxadiazoles.
As shown in Table 4, oxadiazoles with unsubstituted aryl rings were inactive (entries1–5). Interestingly, amide substitution was required for selectivity over TNF-α (R2, in blue). For example, entry 6 (with an –NH2 group) showed no selectivity, whereas entry 7 (with a methyl amide; a confirmed screening hit) was both highly potent towards B cells and was also highly selective. After SAR exploration with 61 other analogs (see Tables 4 and 5), the initial screening hit, CID665654, was therefore selected as one of our probes, ML236 (CID665654). Other aliphatic amides were also potent, but as the size of substitution on the amide increased, various measures of selectivity decreased. For example, an ethyl amide (entry 8) was still very selective over TNF-α, but selectivity decreases from propyl (entry 11), to isopropyl (entry 12), cyclopropyl (entry 13), various butyls (entries 19) as well as larger aliphatic groups and all aromatic groups (entries 20–31). Doubly substituted amides, for example dimethyl (entry 9) and diethyl (entry 10) also showed no or poor selectivity.
Activity was also highly sensitive to substitution around the aromatic ring (R1, in green). 4-t-butyl substitution appeared optimal (as in the probe, entry 7). For example some loss of activity was observed with 4-i-propyl substitution (entry 37) and 4-n-butyl (entry 42), whereas 4-ethyl (entry 32) substitution led to complete loss of activity as well as various methyl, methoxy, ethoxy, and halo substituted compounds (entries 47–58).
Acids and esters in place of amides led to a full loss of selectivity (Table 5). For example the acid derivative of ML263 (entry 60) and methyl ester (entry 61) had no selectivity towards TNF-α (R2, in blue).
A similar SAR strategy was pursued around the oxazole scaffold is depicted in Figure 5. In addition to examining the SAR around the aryl ring (in blue) and amide (in red) as in the oxadiazoles, we also examined the effect of core substitution (in green). The results are summarized in Tables 6–8 below. In general similar SAR trends were observed compared to the oxadiazoles, although there were some differences as noted below.
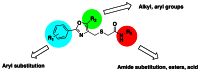
Figure 5
General SAR strategy around oxazole scaffold.
Table 6
SAR Analysis of B-cell Specific Inhibitors of NF-κBActivation: Oxazoles.
Table 7
SAR Analysis of B-cell Specific Inhibitors of NF-κBActivation: Oxazoles.
Table 8
SAR Analysis of B-cell Specific Inhibitors of NF-κBActivation: Oxazoles.
As shown in Table 6, 4-substitution on the aryl ring (R1, in blue) was optimal for activity. An unsubstituted phenyl ring (entry 63) was inactive, but good activity was observed with 4-ethyl (entry 64), 4-thiomethyl (entry 65), and in particular 4-ethoxy (entry 66), which also is the screening hit representative of this oxazole scaffold, was selected as the probe (ML237, CID4174238). All other substitutions weakened the potency or selectivity (see Tables 6, 7 and 8). For example, the 4-Trifluoromethoxy (entry 70), 4-n-propoxy (entry 73) and 4-n-butoxy (entry 74) were also potent and selective, but smaller groups such as 4-trifluoromethyl (entry 69), 4-methoxy (entry 67) and 4-methyl (entry 68) were only weakly active. Also, ortho and meta substituted analogs were inactive, for example 3-ethoxy (entry 71) and 2-ethoxy (entry 72). Lastly, substitution besides methyl at R2 (in green) led to inactive compounds, for example phenyl substitution (entries 75 and 76).
As shown in Table 7, amide substitution greatly impacted activity and selectivity, R1 in green. In general smaller groups such as cyclopropyl (as in entry 66, ML237) were optimal. Methyl (entry 98), isopropyl (entry 77), propyl (entry 78), methoxymethyl (entry 81), cyclopropylmethyl (entry 104) and propargyl (entry 106) all gave compounds with good potency and for the most part selectivity. In contrast to the oxadiazole series, an unsubstituted amide (entry 100) was potent and selective. However, both the carboxylic acid (entry 101) and dimethyl amide (entry 99) either lacked selectivity or potency respectively, which was similar to the oxadiazole series. Aromatic substitution or larger groups on the amide also gave compounds that were either not potent or selective (for example entries 83–89, 91–97, 102 and 105) with the exception of furfuryl (entry 90).
Lastly, as shown in Table 8, we examined a number of combinations of furfuryl at R2 (in green) with additional aromatic groups (R1, in blue). However, a similar SAR trend was seen, with only 4-ethoxy (entry 90, above), 4-propoxy (entry 114) and butoxy (entry 115) giving activity, with other groups being only weakly active (entries 107–111). Also as above, the parent carboxylic acids all lacked selectivity (entries 112–113, 116–118).
3.5. Cellular Activity
All of the primary and selectivity assays are cell based. Specific measures of permeability and toxicity are discussed in section 3.6 below.
3.6. Profiling Assays
The nominated probes were evaluated in a detailed in vitro pharmacology screen as shown in Table 9.
Table 9
Summary of in vitro ADME Properties of B-cell Specific Inhibitors of NF-κBActivation Probes ML236 and ML237.
ML236 and ML237 are moderately soluble in aqueous media at pH 5.0/6.2/7.4.
The PAMPA (Parallel Artificial Membrane Permeability Assay) assay is used as an in vitro model of passive, transcellular permeability. An artificial membrane immobilized on a filter is placed between a donor and acceptor compartment. At the start of the test, drug is introduced in the donor compartment. Following the permeation period, the concentration of drug in the donor and acceptor compartments is measured using UV spectroscopy. Consistent with the predicted LogP (see table 3), ML236 and ML237 are both moderately permeable in this assay.
Plasma Protein Binding is a measure of a drug’s efficiency to bind to the proteins within blood plasma. The less bound a drug is, the more efficiently it can traverse cell membranes or diffuse. Highly plasma protein bound drugs are confined to the vascular space, thereby having a relatively low volume of distribution. In contrast, drugs that remain largely unbound in plasma are generally available for distribution to other organs and tissues. ML236 and ML237 show moderate binding to plasma proteins in mouse and human plasma.
Plasma Stability is a measure of the stability of small molecules and peptides in plasma and is an important parameter, which strongly can influence the in vivo efficacy of a test compound. Drug candidates are exposed in plasma to enzymatic processes (proteinases, esterases), and they can undergo intramolecular rearrangement or bind irreversibly (covalently) to proteins. Both ML236 and ML237 show excellent stability in human and mouse plasma.
The microsomal stability assay is commonly used to rank compounds according to their metabolic stability. This assay addresses the pharmacologic question of how long the parent compound will remain circulating in plasma within the body. ML236 shows excellent stability in human liver microsomes (100% remaining), but poor stability (<1.0% remaining) in mouse liver homogenates, potentially limiting the utility of this probe in in vivo rodent models. ML237 shows poor stability in both human (23% remaining) and mouse (0% remaining) liver homogenates.
ML236 and ML237 show no toxicity (LC50 > 50 μM) toward human hepatocyctes.
Additional Profiling. The probes, ML236 and ML237, were profiled in the NCI-60 panel as well as a DiscoveRx screen of >200 GPCRs, nuclear receptors and kinases. Neither ML236 nor ML 237 showed activity in either the NCI-60 panel or at any of the receptors tested with the exception of the EDG2 receptor (81% and 90% inhibition at 10 μM for ML236 and ML237 respectively). They were submitted to the Psychoactive Drug Screening Program (PDSP) at the University of North Carolina (PDSP, Bryan Roth, PI). These probes are fairly "clean". Of the 42 GPCR receptors tested, ML236 has modest activity against D4 (4.1 μM Ki), and Sigma 1 (8.7 μM Ki), weak activity against 5HT-2B and GABAA (>10 μM Ki), while ML237 has weak activity against only M3 and Sigma1 (>10 μM Ki).
4. Discussion
4.1. Comparison to existing art and how the new probe is an improvement
Previously, a potent (~67 nM) and selective probe (ML029, Pubchem CID2858522) was identified as an inhibitor of the PKC-initiated NF-κB pathway after a battery of 12 secondary and counter-screen assays. However, this probe appeared potent only in the original HEK293 reporter cell line and some types of primary lymphocytes, with little potency in several other tumor cell lines. These previous findings are described in the full Center Probe Report entitled “Chemical inhibitors of antigen receptor-induced NF-κB” (https://mli.nih.gov/mli/?dl_id=454). There are no patents describing inhibitors of NF-κB that are B-cell or T-cell specific.
Because of their activity in B-cells but not T-cells, the newly identified probes are superior to the previously reported pathway-selective NF-κB inhibitors that suppress signaling specifically downstream of PKC without interfering with innate immunity signaling. Cell lineage-selective compounds could serve as powerful research tools for identifying signal transduction components that differ in B-cells versus T-cells. Moreover, compounds that selectively inhibit antigen receptor and PKC-driven NF-κB activation in B-cells but not T-cells have promise for development of therapeutics that could be targeted to cessation of aberrant antibody responses without interfering with cell-mediated immunity (CMI). For example, in diseases such as rheumatoid arthritis and lupus, autoantibody production drives the pathology of these disorders. Selective suppression of humeral limb (B cell mediated) of the adaptive immune response could potentially reduce autoantibody-mediated pathology, without interfering with T-cell-mediated immunity needed for protection from viruses and cancer. Also, in B-cell malignancies (which are far more common than T-cell malignancies), selective interference with the PKC-initiated pathway for NF-κB could be used to inhibit the proliferation and survival of malignant B-cells without interfering with T-cell-mediated immune defenses against viruses. For example, activated B-cell (ABC) type non-Hodgkin’s lymphomas comprise approximately one-quarter of all lymphomas and have hyperactivity of the PKC-initiated pathway for NF-κB activation. Components of this NF-κB pathway, such as MALT1, have been validated by gene knockout and peptide inhibitors to be essential for the growth and survival of these malignant B-cells. Thus, B-cell-selective inhibitors of the PKC-initiated NF-κB pathway could provide a path forward to development of novel anticancer drugs for some common types of malignant lymphoma, where the therapy would be non-immunosuppressive thus affording a superior therapeutic index compared to currently available therapies.
4.2. Mechanism of Action Studies
The mechanism of action (MOA) studies were undertaken and limited in scope to activities modulating secretion of the cytokines IL-2 and IL-8 in Jurkat T-cells. Studies characterizing effects on the modulation of IL-6 and IL-12 in the 697 pre-B-cell lines are on going and will be the subject of a future publication from Dr. Reed’s laboratory.
ML236 did not show any inhibitory activity on the secretion of IL-2 and IL-8 in Jurkat T-cells at concentrations shown to inhibit NF-κB activity in B-cell (IC50B–cells = 0.16 μM) but not T-cells (IC50T–cells = 79.0 μM). Likewise, ML237 did not show any inhibitory activity on the secretion of IL-2 and IL-8 in Jurkat T-cells at concentrations shown to inhibit NF-κB activity in B-cell (IC50B–cells = 0.19 μM) but not T-cells (IC50T–cells = 79.0 μM).
4.3. Planned Future Studies
Currently, our immediate future plans are to establish ELISA assays to distinguish the ability of ML236 and ML237 to inhibit IL-6 and IL-12 secretion in B-cells. In parallel the activity of the compounds will be tested against primary cultured human lymphocytes and ABC-type lymphoma cell lines. Downstream steps in the signaling transduction cascade initiated by antigen receptors (and PKC) will be interrogated as described for the prior probe compound ML025 (Shi, R, et al. ACS Chemical Biology 2010), including proteolytic processing of Bcl-10 and A20 (substrates of MALT1 protease), formation of the CARMA/Bcl-10/MALT (CBM) complex (assessed by co-immunoprecipitation experiments), phosphorylation of CARMA (determined by phosphospecific antibody immunoblotting), and ubiquitination of TRAF6 and NEMO/IKK-gamma. The goal is to elucidate the step in the pathway where the compound blocks the signaling cascade, thus providing insights into the possible cellular mechanisms of action.
5. References
- 1.
- McAllister-Lucas LM, Inohara N, Lucas PC, Ruland J, Benito A, Li Q, Chen S, Chen FF, Yamaoka S, Verma IM, et al. Bimp1, a MAGUK family member linking protein kinase C activation to Bcl10-mediated NF-kappaB induction. J Biol Chem. 2001;276:30589–30597. [PubMed: 11387339]
- 2.
- Ruefli-Brasse AA, French DM, Dixit VM. Regulation of NFkappaB-dependent lymphocyte activation and development by paracaspase. Science. 2003;302:1581–1584. [PubMed: 14576442]
- 3.
- Ruland J, Duncan GS, Elia A, del Barco Barrantes I, Nguyen L, Plyte S, Millar DG, Bouchard D, Wakeham A, Ohashi PS, Mak TW. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-kappaB and neural tube closure. Cell. 2001;104:33–42. [PubMed: 11163238]
- 4.
- Thome M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004;4:348–359. [PubMed: 15122200]
- 5.
- Zhao KW, Li X, Zhao Q, Huang Y, Li D, Peng ZG, Shen WZ, Zhao J, Zhou Q, Chen Z, et al. Protein kinase Cdelta mediates retinoic acid and phorbol myristate acetate-induced phospholipid scramblase 1 gene expression: its role in leukemic cell differentiation. Blood. 2004;104:3731–3738. [PubMed: 15308560]
- 6.
- DiMauro Erin F, Buchanan John L, Cheng Alan, Emkey Renee, Hitchcock Stephen A, Huang Liyue, Huang Ming Y, Janosky Brett, Lee Josie H, Li Xingwen, Martin Matthew W, Tomlinson Susan A, White Ryan D, Zheng Xiao Mei, Patel Vinod F, Fremeau Robert T Jr. Structural modifications of N-arylamide oxadiazoles: Identification of N-arylpiperidine oxadiazoles as potent and selective agonists of CB2. Bioorg Med Chem Lett. 2008;18:4267–4274. [PubMed: 18640038]
6. Supplementary Information
6.1. Assay Details
NF-κB B-Cell Primary Assay and Dose Response Protocol
Assay materials
- 697 NF-kB-Luc cell line obtained from Dr. Reed’s lab
- RPMI phenol red (−), Mediatech Car # 17105CV
- Fetal Bovine Serum, Hyclone cat # SH30396.03HI
- L-glutamine. Omega Scientific cat # GS-60
- Na-pyruvate, Sigma cat # S8636-100M
- Penn/Strep, Omega Scientific cat # PS-20
- PMA, Calbiochem cat # 524400
- Ionomycin, Calbiochem cat # 407950
- SteadyGlo, Promega cat # E2550
Day 1 Procedure
- Harvest 697 NF-κB-Luc at 2,000,000 cells/mL density. Spin, re-suspend in assay media, count.
- Seed 15000 cells/well in 3 μL/well 697 NF-kB-Luc to full plate- Aurora 1536 low base white plate # 00029846 plate or non-TC-treated equivalent.
- Plate with Kalypsys dispenser
- Spin down plates @ 500 RPM for 1 min.
- Dispense 10 nL 100% DMSO compounds (col 5–48) & 10nL 100% DMSO controls (col 1–4) to plates with Echo 555 (Labcyte) acoustic dispenser (For dose response assays, the appropriate volume and concentration of 10 mM test compounds is transferred to assay plate to achieve desired final assay concentration and range. Wells are backfilled with DMSO to equalize DMSO concentrations between wells)
- Incubate plate for 1 hour in 37°C 5% CO2.
- Add 2μL PMA/Ionomycin working dilution (6.5 ng/mL PMA/3.25 ng/mL Ionomycin) in assay media to col 3–48 and no agonist media to col 1 and 2 with Kalypsys dispenser.
- Spin down plates @ 1000 RPM for 1 min.
- Lid plates with Kalypsys plate lids.
- Incubate plate overnight (16 hours) in 37°C 5% CO2 incubator.
Day 2 Procedure
- Remove lid and incubate plate for 10 min in at room temp.
- Add 3 μL/well SteadyGlo with Kalypsys dispenser
- Spin plates @ 2000 RPM for 2 min, lid plate and incubate for 10 min at room temp.
- Read luminescence on Envision Ultra sensitive Luminescent protocol for white Aurora 1536 low base white plate # 00029846
NF-κB T-Cell Primary Assay and Dose Response Protocol
Assay materials
- Jurkat NF-kB cell line obtained from Dr. Reed’s lab
- RPMI phenol red (−), Mediatech Car # 17105CV
- Fetal Bovine Serum, Hyclone cat # SH30396.03HI
- L-glutamine. Omega Scientific cat # GS-60
- Na-pyruvate, Sigma cat # S8636-100M
- Penn/Strep, Omega Scientific cat # PS-20
- PMA, Calbiochem cat # 524400
- Ionomycin, Calbiochem cat # 407950
- SteadyGlo, Promega cat # E2550
Day 1 Procedure
- Harvest Jurkat NF-κB-Luc at 1000,000 cells/mL density. Spin, resuspend in assay media, count.
- Seed 8000 cells/well in 3 μL/well to full plate Jurkat NF-κB -Luc to white Aurora 1536 low base white plate # 00029846 plate or non-TC-treated equivalent.
- Plate with Kalypsys dispenser
- Spin down plates @ 500 RPM for 1 min.
- Dispense 10nL 100% DMSO compounds (col 5–48) & 10 nL 100% DMSO controls (col 1–4) to plates with Echo 555 (Labcyte) acoustic dispenser (For dose response assays, the appropriate volume and concentration of 10 mM test compounds is transferred to assay plate to achieve desired final assay concentration and range. Wells are backfilled with DMSO to equalize DMSO concentrations between wells)
- Incubate plate for 1 hour in 37°C 5% CO2.
- Add 2μL PMA/Ionomycin working dilution (7.5 ng/mL PMA/3.75 ng/mL Ionomycin) in assay media to col 3–48 and no agonist media to col 1 and 2 with Kalypsys dispenser.
- Spin down plates @ 1000 RPM for 1 min.
- Lid plates with Kalypsys plate lids.
- Incubate plate overnight (16 hours) in 37°C 5% CO2 incubator.
Day 2 Procedure
- Remove lid and incubate plate for 10 min in at room temp.
- Add 3μL/well SteadyGlo with Kalypsys dispenser
- Spin plates @ 2000 RPM for 2 min, lid plate and incubate for 10 min at room temp.
- Read luminescence on Envision Ultra sensitive Luminescent protocol for white Aurora 1536 low base white plate # 00029846
TNF-a NF-κB induction Assay: Dose Response
Assay materials
- HEK-293-T NF-κB-Luc cell line obtained from Dr. Reed’s lab’s laboratory
- Assay Media: DMEM with no phenol red (Gibco 31053 or Hyclone SH30585.02), 2mM L-glutamine (omega sci GS-60 200mM), 1mM Na-pyruvate (Sigma S8636-100ML 100mM), 50IU/ml Penn/Strep(omega scientific PS-20 5000 IU/mL)
- Recombinant Human TNF-alpha, CF, (R&D Systems 210-TA-010/CF)
- SteadyGlo (Promega E2550)
- Assay plate: 1536-well, TC-treated, black plate (Corning 3893)
Day 1 Procedure
- Harvest HEK-293-T NF-κB-Luc at 100% confluency
- Make cells suspension at 1X for final seeding density of 6000 cells/well in 5μL well volume.
- Add TNF-a to 1X cell suspension at 0.1 ng/mL.
- Dispense 6000 cells/well in 5 μL of assay media into col. 5–48 of assay plate
- Dispense 5 μL/well of assay media into col. 1–4 of assay plate
- Spin down plates at 1000 rpm for 1 min in an Eppendorf 5810 centrifuge
- With LabCyte Echo, dispense varying volumes of test compounds in DMSO into assay plate wells (Col. 5–44) to achieve appropriate dose response concentrations and range. Backfill test compound wells with DMSO to equilibrate DMSO concentrations. Transfer equal volumes of DMSO to control wells (Col. 1–4, 45–48). Maximum concentration of DMSO should be 1%.
- Lid Plates. Sandwich 4 plates between 2 lidded 384 plates filled with H2O
- Wrap plates securely in single layer of Plastic Wrap (Saran Wrap PVDC version).
- Incubate overnight (14 hours) in 37°C 5% CO2 incubator
Day 2 Procedure
- Add 3 μL/well SteadyGlo to each well with Multidrop.
- Spin plates at 1000 rpm for 1 minute on Eppendorf 5810 centrifuge
- Read luminescence on Perkin-Elmer EnVision
Cytotoxicity Assay: Dose Response
Assay materials
- HEK-293-T NF-κB-Luc cell line obtained from Dr. Reed’s lab’s laboratory
- Assay Media: DMEM with no phenol red (Gibco 31053 or Hyclone SH30585.02), 2mM L-glutamine (omega sci GS-60 200mM), 1mM Na-pyruvate (Sigma S8636-100ML 100mM), 50IU/ml Penn/Strep(omega scientific PS-20 5000 IU/mL)
- Resazurin (Sigma-Aldrich R7017)
- Assay plate: 1536-well, TC-treated, black plate (Corning 3893)
Day 1 Procedure
- Harvest HEK-293-T NF-κB-Luc at 100% confluency
- Make cells suspension at 1X for final seeding density of 6000 cells/well in 5μL well volume.
- Dispense 6000 cells/well in 5 μL of assay media into col. 5–48 of assay plate
- Dispense 5 μL/well of assay media into col. 1–4 of assay plate as positive control.
- Spin down plates at 1000 rpm for 1 min in an Eppendorf 5810 centrifuge
- With LabCyte Echo, dispense varying volumes of test compounds in DMSO into assay plate wells (Col. 5–44) to achieve appropriate dose response concentrations and range. Backfill test compound wells with DMSO to equilibrate DMSO concentrations. Transfer equal volumes of DMSO to control wells (Col. 1–4, 44–48). Maximum concentration of DMSO should be 1%.
- Lid Plates. Sandwich 4 plates between 2 lidded 384 plates filled with H2O
- Wrap plates securely in single layer of Plastic Wrap (Saran Wrap PVDC version).
- Incubate overnight (14 hours) in 37°C 5% CO2 incubator
Day 2 Procedure
- Dispense 20nL of 2mM Resazarin in DMSO (FAC is 8uM) with LabCyte Echo to all wells of assay plate
- Lid Plates. Sandwich 4 plates between 2 lidded 384 plates filled with H2O
- Wrap plates securely in single layer of Plastic Wrap (Saran Wrap PVDC version).
- Incubate for 1 hour in 37°C 5% CO2 incubator
- Top read Fluorescence Intensity (Ex. 531, Em. 595nm) on Perkin-Elmer EnVision
6.2. Critical Path Flowchart for NF-κB B-cell Inhibitors
The critical path for the selection of the probe and intended further studies for the probe are shown below. The actual assay was conducted as comparative screen for each cell type and the differential activities were used to define whether a compound was B-, T- or B & T-cell dual selective.
6.3. Supplementary Information for Probe Characterization
- PubChem BioAssay for Chemical ProbePubChem BioAssay records reporting screening data for the development of the chemical probe(s) described in this book chapter
- PubChem SubstanceRelated PubChem Substances
- PubMedLinks to PubMed
- Salvia miltiorrhiza polysaccharide activates T Lymphocytes of cancer patients through activation of TLRs mediated -MAPK and -NF-κB signaling pathways.[J Ethnopharmacol. 2017]Salvia miltiorrhiza polysaccharide activates T Lymphocytes of cancer patients through activation of TLRs mediated -MAPK and -NF-κB signaling pathways.Chen Y, Li H, Li M, Niu S, Wang J, Shao H, Li T, Wang H. J Ethnopharmacol. 2017 Mar 22; 200:165-173. Epub 2017 Feb 21.
- Novel non-peptide small molecules preventing IKKβ/NEMO association inhibit NF-κB activation in LPS-stimulated J774 macrophages.[Biochem Pharmacol. 2016]Novel non-peptide small molecules preventing IKKβ/NEMO association inhibit NF-κB activation in LPS-stimulated J774 macrophages.De Falco F, Di Giovanni C, Cerchia C, De Stefano D, Capuozzo A, Irace C, Iuvone T, Santamaria R, Carnuccio R, Lavecchia A. Biochem Pharmacol. 2016 Mar 15; 104:83-94. Epub 2016 Jan 14.
- Chemical biology strategy reveals pathway-selective inhibitor of NF-kappaB activation induced by protein kinase C.[ACS Chem Biol. 2010]Chemical biology strategy reveals pathway-selective inhibitor of NF-kappaB activation induced by protein kinase C.Shi R, Re D, Dudl E, Cuddy M, Okolotowicz KJ, Dahl R, Su Y, Hurder A, Kitada S, Peddibhotla S, et al. ACS Chem Biol. 2010 Mar 19; 5(3):287-99.
- Review Anti-apoptotic action of API2-MALT1 fusion protein involved in t(11;18)(q21;q21) MALT lymphoma.[Apoptosis. 2005]Review Anti-apoptotic action of API2-MALT1 fusion protein involved in t(11;18)(q21;q21) MALT lymphoma.Hosokawa Y. Apoptosis. 2005 Jan; 10(1):25-34.
- Review NF-kappaB signaling in lymphocytes: a new cast of characters.[J Cell Sci. 2004]Review NF-kappaB signaling in lymphocytes: a new cast of characters.Lucas PC, McAllister-Lucas LM, Nunez G. J Cell Sci. 2004 Jan 1; 117(Pt 1):31-9.
- B-cell Specific Inhibitors of NF-κB Activation - Probe Reports from the NIH Mole...B-cell Specific Inhibitors of NF-κB Activation - Probe Reports from the NIH Molecular Libraries Program
Your browsing activity is empty.
Activity recording is turned off.
See more...