NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
A high-throughput screen (HTS) of the Molecular Libraries Probe Centers Network (MLPCN) library was performed using a thallium influx assay in order to identify inhibitors of potassium voltage-gated channel, KQT-like subfamily, member 2 (KCNQ2) channels. Structure activity relationship (SAR) studies of active compounds yielded ML252, a potent (IC50 = 69 nM) inhibitor of KCNQ2 channels in electrophysiological assays. ML252 displayed more than forty-fold selectivity for blocking KCNQ2 channels compared with KCNQ1 channels. SAR studies revealed a site on ML252 at which small structural changes caused a functional shift from antagonist to agonist activity, suggesting that ML252 interacted with a critical site for controlling gating of KCNQ2 channels. ML252 represents a novel and potent inhibitor of KCNQ2 channels with a selectivity profile that enables use of the probe for investigating the role of KCNQ2 channels in neuronal function.
Assigned Assay Grant #: 1 R03 DA027716-01
Screening Center Name & PI: Johns Hopkins Ion Channel Center, Min Li
Chemistry Center Name & PI: Vanderbilt Specialized Chemistry Center for Accelerated Probe Development, Craig W. Lindsley
Assay Submitter & Institution: Min Li, Johns Hopkins University School of Medicine
PubChem Summary Bioassay Identifier (AID): 2262
Probe Structure & Characteristics
| CID/ML# | Target Name | IC50 (nM) [SID, AID] | Anti-target Name(s) | IC50 (μM) [SID, AID] | Fold Selective | Secondary Assay(s) Name: IC50/EC50 (nM) [SID, AID] |
|---|---|---|---|---|---|---|
| CID 50985951 / ML252 | KCNQ2 | 69 [SID 116933760, AID 588426] | KCNQ1 | 2.92 [SID 116933760, AID 588425] | >40 |
Recommendations for Scientific Use of the Probe
KCNQ2 channels play a role in controlling excitability and regulating transmitter release from some neuronal populations. Studies with nonselective KCNQ channel inhibitors have shown a role for these channels in regulating acetylcholine release [1], suggesting a role for KCNQ channel inhibitors as a possible treatment paradigm for Alzheimer’s disease [2]. ML252 provides a potent and novel chemical probe with a selectivity profile that differs from previously described KCNQ2 channel inhibitors. ML252 may be used to investigate the role of KCNQ2 channels in regulating membrane excitability and neurotransmitter release using in vitro assay systems. This information will be useful to investigators studying the role of potassium channels in neurotransmitter release and cell excitability mechanisms.
2. Materials and Methods
- CHO-KCNQ2 cell line
- Parental CHO cell line
- CHO-KCNQ1 cell line
- CHO-KCNQ1/KCNE1 cell line
- CHO-KCNQ2/3 cell line
- CHO-KCNQ4 cell line
- FluxOR dye kit (Invitrogen, F10017)
- BD Biocoat, Poly-D-Lysine coated, black/clear bottom 384-well assay plates (Fisher, 356936)
- Hamamatsu FDSS 6000 fluorescent plate reader
- Population Patch Clamp patch plates from Molecular Devi
2.1. Assays
- Summary AID 2262: Summary of probe development for inhibitors of KCNQ2 potassium channel
- AID 2156: Primary cell-based high-throughput screening assay for identification of compounds that inhibit KCNQ2 potassium channels
- AID 493025: Confirmatory screen for compounds that inhibit KCNQ2 potassium channels
- AID 493029: Counter screen against parental CHO cells for compounds that inhibit KCNQ2 potassium channels
- AID 588531: Confirmatory screen for compounds that inhibit KCNQ2 potassium channels using automated patch clamp
- AID 588637: Dose response assay for compounds that inhibit KCNQ2 potassium channels on automated electrophysiological assay II
- AID 493026: Specificity screen against KCNQ1 for compounds that inhibit KCNQ2 potassium channels
- AID 588426: SAR analysis for compounds that inhibit KCNQ2 potassium channels on automated electrophysiological assay, CRC2
- AID 588425: Dose response assay of SAR compounds for the identification of selective inhibitors of KCNQ2 potassium channels in the KCNQ1 expressing cells on automated patch clamp
- AID 504837: SAR analysis for compounds that inhibit KCNQ2 potassium channels on automated electrophysiological assay
- AID 504839: Dose response assay for compounds that inhibit KCNQ2 potassium channels on automated electrophysiological assay
- AID 588635: SAR-Confirmatory assay to confirm a potent KCNQ2 inhibitor using manual electrophysiology
2.2. Probe Chemical Characterization
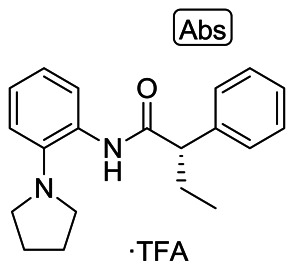
Probe compound ML252 (CID: 50985951, SID: 116933760) was prepared according to the scheme in Figure 1 and provided the following characterization data: LCMS (>98%) m/z = 309.3 [M + H]+ (0.66 min retention time, 215 and 254 nm). 1H NMR (400 MHz, DMSO-d6) δ 10.10 (br s, 1H), 7.52–7.41 (m, 3H), 7.41 – 7.30 (m, 3H), 7.31–7.10 (m, 3H), 3.76 (t, J = 7.5 Hz, 1H), 3.72–3.42 (m, 1H), 3.84 (br s, 4H), 2.20–2.02 (m, 1H), 1.88 (br s, 4H), 1.81–1.66 (m, 1H), 0.89 (t, J = 7.3 Hz, 3H). Chiral HPLC: 94.2% (S) RT = 3.97 min: 5.75% (R) RT = 4.29 min; 88.5% ee.
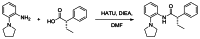
Figure 1
Synthesis of ML252.
Solubility: Solubility in PBS was determined to be 14 μM, which is more than 200-fold higher than the EC50 for KCNQ2 channel inhibition.
Stability: Stability was determined for ML252 at 23°C in PBS (no antioxidants or other protectorants and DMSO concentration below 0.1%; see appendix 4). After 48 hours, 66% of the initial concentration of ML252 remained (Table 1; Figure 2).
Table 1
Stability assessment in PBS (Absorption Systems).
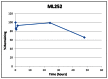
Figure 2
Stability assessment in PBS (Absorption Systems).
Compounds added to the SMR collection (MLS#s): MLS003871689 (ML252, CID 50985951, 20.0 mg); MLS003871690 (CID 53313402, 5.3 mg); MLS003871691 (CID 53313363, 5.0 mg); MLS003871692 (CID 53313387, 5.4 mg); MLS003871693 (CID 53313379, 5.0 mg); MLS003871694 (CID 53313381, 6.1 mg)
2.3. Probe Preparation
(S)-2-Phenyl-N-(2-(pyrrolidin-1-yl)phenyl)butanamide (ML252, CID: 50985951). To a stirred solution of (S)-(−)-2-phenylbutyric acid (50 mg, 0.30 mmol), HATU (116 mg, 0.304 mmol) in DMF (1.5 mL) was added ethyldiisopropylamine (79 μL, 0.46 mmol) followed by 2-(1-pyrrolidinyl)aniline (49 mg, 0.30 mmol) and the reaction mixture was stirred at room temperature overnight. The reaction was diluted with water (1.5 mL) and extracted with EtOAc (2 × 2 mL). The organic extracts were combined, concentrated and the residue was purified by RP prepHPLC eluting with 10 to 90% CH3CN/H2O (0.1% TFA) to give the product as the TFA salt (60 mg, 57%).
3. Results
3.1. Dose Response Curves for Probe
ML252 displayed potent block of KCNQ2 channels in automated electrophysiology experiments using IonWorks instruments. The protocol used to evaluate block of KCNQ2 channels is shown in Fig. 3A. Currents from a representative well (Fig. 3A) were significantly inhibited by 0.3 μM ML252. In dose-response experiments (Fig. 3B), such as the one shown in Fig. 3B, ML252 displayed an average IC50 value of 69 ± 6 nM (mean ± SD; n=6) and produced nearly complete block at concentrations above 1 μM.
3.2. Cellular Activity
The primary HTS assay and all secondary assays are cell-based assays, indicating that ML252 can gain access to its molecular target when applied to cells. The compound did not exhibit acute toxicity in cell-based assays at concentrations up to 30 μM.
3.3. Profiling Assays
ML252 (CID 50985951) has been tested in 301 assays performed within the MLPCN network and was active in only four assays that were not dependent on KCNQ2. ML252 showed >300-fold selectivity versus these assays (Table 2).
Table 2
Selectivity profile from PubChem.
To more fully characterize this novel KCNQ2 inhibitor, ML252 was tested on Ricerca’s (formerly MDS Pharma’s) Lead Profiling Screen (binding assay panel of 68 GPCRs, ion channels and transporters screened at 10 μM), and was found to bind with only 1 of the 68 assays conducted (no inhibition of radio ligand binding > 50% at 10 μM) (Table 3). Included in the Ricerca screening panel are a number of ion channels (Calcium Channel, L-Type and N-Type; Potassium channel [KATP]; Potassium channel [hERG]) showing the selectivity profile for ML252.
Table 3
Ricerca Profiling of ML252 (CID 50985951).
ML252 selectivity for blocking KCNQ channel family members was examined using automated electrophysiology as described in AID588426 (Table 5). ML252 displayed approximately 40-fold selectivity for block of KCNQ2 channels compared with KCNQ1 channels (IC50 = 2.92 μM). The heteromultimeric combination of KCNQ1 with KCNE1 forms the Iks current [3], which is a key repolarization component in cardiac myocytes. ML252 afforded greater than 100-fold selectivity for block of KCNQ2 compared with KCNQ1/E1 (IC50 = 8.12 μM). ML252 displayed little selectivity for block of KCNQ2/Q3 heteromultimeric channels (IC50 = 0.12 μM) and modest selectivity for block of KCNQ4 channels (IC50 = 0.20 μM).
4. Discussion
4.1. Comparison to Existing Art and How the New Probe is an Improvement
The prior art consists of three known compounds (XE991, Linopridine, and UCL2077). Each of these compounds show mid-nanomolar to micromolar activity against KCNQ2 based on published values (Table 4, right column) [4][5]. The newly reported probe (ML252) is >10-fold more potent at KCNQ2 than known compounds based on literature values, which were generated using different experimental approaches. In order to directly compare ML252 with previously described KCNQ2 blockers using a common assay system, we evaluated these compounds using IonWorks automated electrophysiology (AID588426) for block of KCNQ2 and other KCNQ family members (Table 4). ML252 affords equivalent or higher potency along with increased selectivity against blocking KCNQ1 channels. ML252 displayed approximately 40-fold selectivity for block of KCNQ2 channels compared with KCNQ1 channels. In contrast, XE991 and linopiridine were also potent blockers of KCNQ2 channels, but displayed reduced selectivity versus KCNQ1 channels. The heteromultimeric combination of KCNQ1 with KCNE1 forms the Iks current [3], which is a key repolarization component in cardiac myocytes. ML252 afforded greater than 100-fold selectivity for block of KCNQ2 compared with KCNQ1/E1, while XE991 and linopiridine displayed 26-fold and 3-fold selectivity, respectively. Linopiridine and ML252 blocked KCNQ2 channels at submicromolar concentrations, with less than two-fold selectivity for blocking KCNQ2/Q3 heteromultimeric channels, while XE991 also blocked KCNQ2 channels, but displayed five-fold selectivity against KCNQ2/Q3 channels.
Table 4
Selectivity table showing IC50 (μM) values for ML252 and previously identified KCNQ2 channel blockers for inhibition of a panel of KCNQ channels measured using IonWorks electrophysiology.
Unlike previously described KCNQ2 inhibitors, ML252 displays striking structure-activity properties, in which small structural changes in ML252 can lead to a functional switch between antagonist and agonist behavior. These properties may allow ML252 to serve as a useful tool for molecular studies of KCNQ2 channel gating.
5. References
- 1.
- Earl RA, et al. 2-Fluoro-4-pyridinylmethyl analogues of linopirdine as orally active acetylcholine release-enhancing agents with good efficacy and duration of action. J Med Chem. 1998;41(23):4615–22. [PubMed: 9804701]
- 2.
- Chorvat RJ, Zaczek R, Brown BS. Ion channel modulators that enhance acetylcholine release: potential therapies for Alzheimer's disease. Expert Opin Investig Drugs. 1998;7(4):499–518. [PubMed: 15991988]
- 3.
- Sanguinetti MC, et al. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384(6604):80–3. [PubMed: 8900283]
- 4.
- Wang HS, et al. KCNQ2 and KCNQ3 potassium channel subunits: molecular correlates of the M-channel. Science. 1998;282(5395):1890–3. [PubMed: 9836639]
- 5.
- Soh H, Tzingounis AV. The specific slow afterhyperpolarization inhibitor UCL2077 is a subtype-selective blocker of the epilepsy associated KCNQ channels. Mol Pharmacol. 78(6):1088–95. [PMC free article: PMC2993466] [PubMed: 20843955]
- PMCPubMed Central citations
- PubChem BioAssay for Chemical ProbePubChem BioAssay records reporting screening data for the development of the chemical probe(s) described in this book chapter
- PubChem SubstanceRelated PubChem Substances
- PubMedLinks to PubMed
- Review A small molecule activator of KCNQ2 and KCNQ4 channels.[Probe Reports from the NIH Mol...]Review A small molecule activator of KCNQ2 and KCNQ4 channels.Yu H, Wu M, Hopkins C, Engers J, Townsend S, Lindsley C, McManus OB, Li M. Probe Reports from the NIH Molecular Libraries Program. 2010
- Review ML365: Development of Bis-Amides as Selective Inhibitors of the KCNK3/TASK1 Two Pore Potassium Channel.[Probe Reports from the NIH Mol...]Review ML365: Development of Bis-Amides as Selective Inhibitors of the KCNK3/TASK1 Two Pore Potassium Channel.Zou B, Flaherty DP, Simpson DS, Maki BE, Miller MR, Shi J, Wu M, McManus OB, Golden JE, Aubé J, et al. Probe Reports from the NIH Molecular Libraries Program. 2010
- Discovery of a series of 2-phenyl-N-(2-(pyrrolidin-1-yl)phenyl)acetamides as novel molecular switches that modulate modes of K(v)7.2 (KCNQ2) channel pharmacology: identification of (S)-2-phenyl-N-(2-(pyrrolidin-1-yl)phenyl)butanamide (ML252) as a potent, brain penetrant K(v)7.2 channel inhibitor.[J Med Chem. 2012]Discovery of a series of 2-phenyl-N-(2-(pyrrolidin-1-yl)phenyl)acetamides as novel molecular switches that modulate modes of K(v)7.2 (KCNQ2) channel pharmacology: identification of (S)-2-phenyl-N-(2-(pyrrolidin-1-yl)phenyl)butanamide (ML252) as a potent, brain penetrant K(v)7.2 channel inhibitor.Cheung YY, Yu H, Xu K, Zou B, Wu M, McManus OB, Li M, Lindsley CW, Hopkins CR. J Med Chem. 2012 Aug 9; 55(15):6975-9. Epub 2012 Jul 26.
- Review Identification of a novel, small molecule activator of KCNQ1 channels.[Probe Reports from the NIH Mol...]Review Identification of a novel, small molecule activator of KCNQ1 channels.Yu H, Lin Z, Xu K, Huang X, Long S, Wu M, McManus OB, Le Engers J, Mattmann ME, Engers DW, et al. Probe Reports from the NIH Molecular Libraries Program. 2010
- Review Development of a Selective Chemical Inhibitor for the Two-Pore Potassium Channel, KCNK9.[Probe Reports from the NIH Mol...]Review Development of a Selective Chemical Inhibitor for the Two-Pore Potassium Channel, KCNK9.Miller MR, Zou B, Shi J, Flaherty DP, Simpson DS, Yao T, Maki BE, Day VW, Douglas JT, Wu M, et al. Probe Reports from the NIH Molecular Libraries Program. 2010
- Identification of a novel, small molecule inhibitor of KCNQ2 channels - Probe Re...Identification of a novel, small molecule inhibitor of KCNQ2 channels - Probe Reports from the NIH Molecular Libraries Program
Your browsing activity is empty.
Activity recording is turned off.
See more...